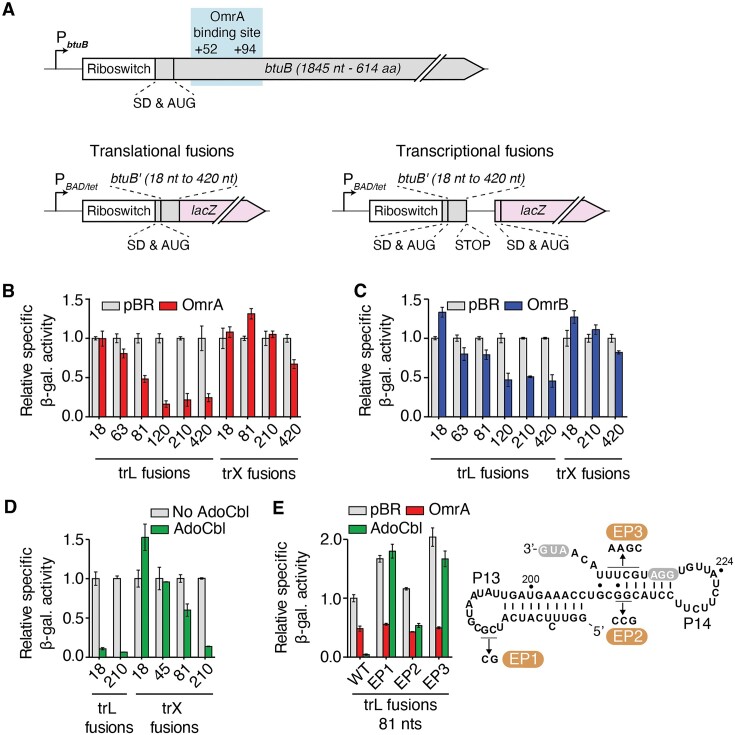
Figure 2.
sRNAs and riboswitch regulation rely on different regions of the btuB mRNA. (A) Schematics showing the btuB gene with the location of the OmrA binding site (top) and translational (bottom left) and transcriptional (bottom right) fusions containing various lengths of btuB coding region (btuB'). An arabinose inducible (PBAD) or a constitutive (PLtetO-1) promoter was used to express the constructs. The Shine-Dalgarno (SD) and AUG are indicated. (B–D) β-galactosidase assays of translational (trL) BtuB-LacZ and transcriptional (trX) btuB-lacZ fusions in the presence of OmrA (B), OmrB (C) or AdoCbl (D). The number of nucleotides of the btuB CDS is indicated for each construct. Values were normalized to the activity obtained in the absence of sRNA (pBR, empty vector) or without AdoCbl. The average values and the standard deviations were obtained from three independent experiments. (E) Beta-galactosidase assays of selected btuB mutants destabilizing the stems P13 and P14. Experiments were performed using the BtuB81-LacZ translational fusion in the presence of OmrA or AdoCbl. The predicted secondary structure of the region encompassing stems P13 and P14 is shown to the right, and the EP1, EP2 and EP3 mutations are indicated. The SD sequence (GGA) and the AUG are white in gray.