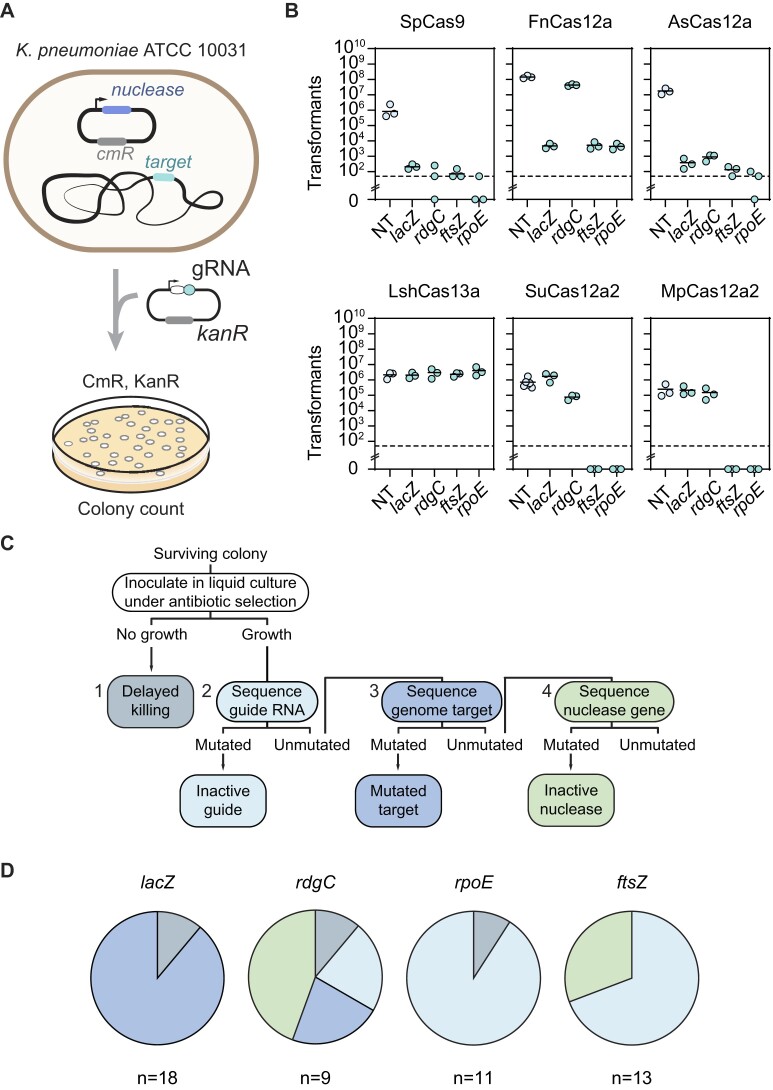
Figure 1.
DNA-targeting nucleases outperform RNA-targeting nucleases as CRISPR antimicrobials for the selected targets, with AsCas12a exhibiting high antimicrobial activity against K. pneumoniae that drives infrequent but variable escape. (A) Experimental setup for the transformation-based targeting assay. (B) Drop in transformants following genome targeting with different Cas single-effector nucleases. Each dot represents a biological replicate obtained by preparing electrocompetent cells from a separate colony. ftsZ and rpoE: essential genes. rdgC and lacZ: non-essential genes. The dashed lines represent the limit-of-detection for colony numbers. The bars represent average values and are absent for samples in which at least one replicate had no colonies. (C) Flowchart of procedure to identify escape mutants. The mutations were screened following the indicated order until an escape mutation was identified. (D) Pie chart of the modes of escape from targeting for the different gRNAs. n indicates the number of screened colonies per condition.