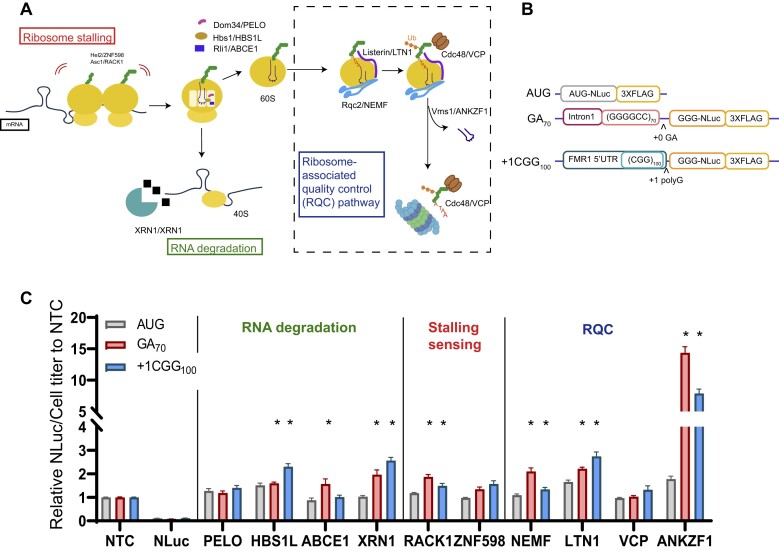
Figure 1.
NEMF, LTN1, and ANKZF1 act as genetic modifiers of RAN translation from both G4C2 and CGG repeats. (A) Stalled or collided ribosomes are sensed by ZNF598 and RACK1 to separate 80S ribosome to 60S and 40S. The 40S subunit with truncated mRNA is then released by PELO in concert with HBS1L and ABCE1 and subsequently degraded by XRN1. NEMF CAT-tails partially generated peptides within the 60S subunit, allowing for degradation by the proteasome through a process that involves ubiquitination by LTN1 and VCP. ANKZF1 removes the tRNA, allowing for ribosomal recycling. Schematic adapted with permission from (63). (B) Schematic of transiently expressed nano-luciferase (NLuc) reporters used to assess RAN translation product abundance. Either the first intron of C9orf72 including 70 GGGGCC (G4C2) repeats or the 5′UTR of FMR1 containing 100 CGG repeats were placed 5′ to a modified NLuc with the AUG initiator codon mutated to GGG. (C) Results from a targeted screen of key RQC pathway factors. The relative expression of NLuc was normalized to cell titer. These normalized values were then expressed as a fold change compared to a non-targeting siRNA control (NTC). The effect of transient transfection of each siRNA or luciferase reporter on cell titer is shown in Supplemental Figure S1 (Supplementary Figure S1). Data represent mean with error bars ± SEM of 6 biological replicates from at least 2 independent experiments. *P< 0.05; one-way ANOVA with Dunnett's multiple comparison test compared to NTC.