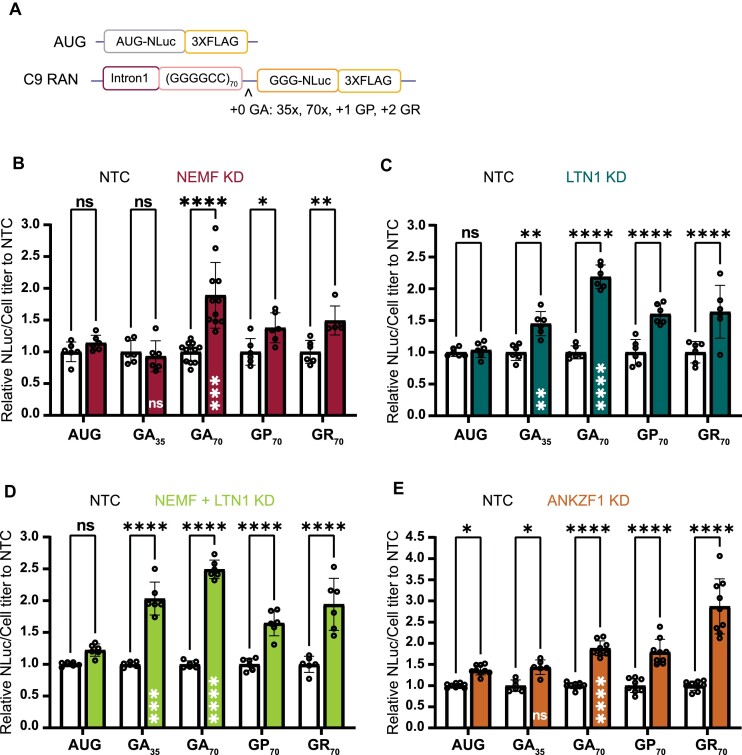
Figure 2.
Depletion of NEMF, LTN1, and ANKZF1 enhances RAN translation in a repeat length-dependent manner and across all reading frames. (A) Schematic of C9orf72 RAN G4C2 repeat length and reading frame reporters. Single nucleotide insertions to shift the reading frame and repeat contractions allowed for measurement of products from all potential reading frames and across 2 repeat sizes. (B–E) Luciferase assays after NEMF, LTN1, both NEMF and LTN1 or ANKZF1 depletion. All graphs show mean with error bars ± SD. Each N is shown as an open circle (n = 6–9/group across at least two independent experiments). Asterisks above each bar are comparisons of expression between NTC and gene(s) knockdown. ns = not significant; *P< 0.05; **P≤ 0.01; ***P≤ 0.001; ****P≤ 0.0001, as determined with two-way ANOVA with Sidak's multiple comparison test. Asterisks placed inside each bar are comparisons between AUG-driven no-repeat control and different repeat lengths of the GA frame of cells treated with gene(s) knockdown. ns = not significant; **P≤ 0.01; ***P≤ 0.001; ****P≤ 0.0001, represent unpaired t-test.