Fig. 3 |. Choosing genetically encoded neurotransmitter or neuromodulator indicators for experiments.
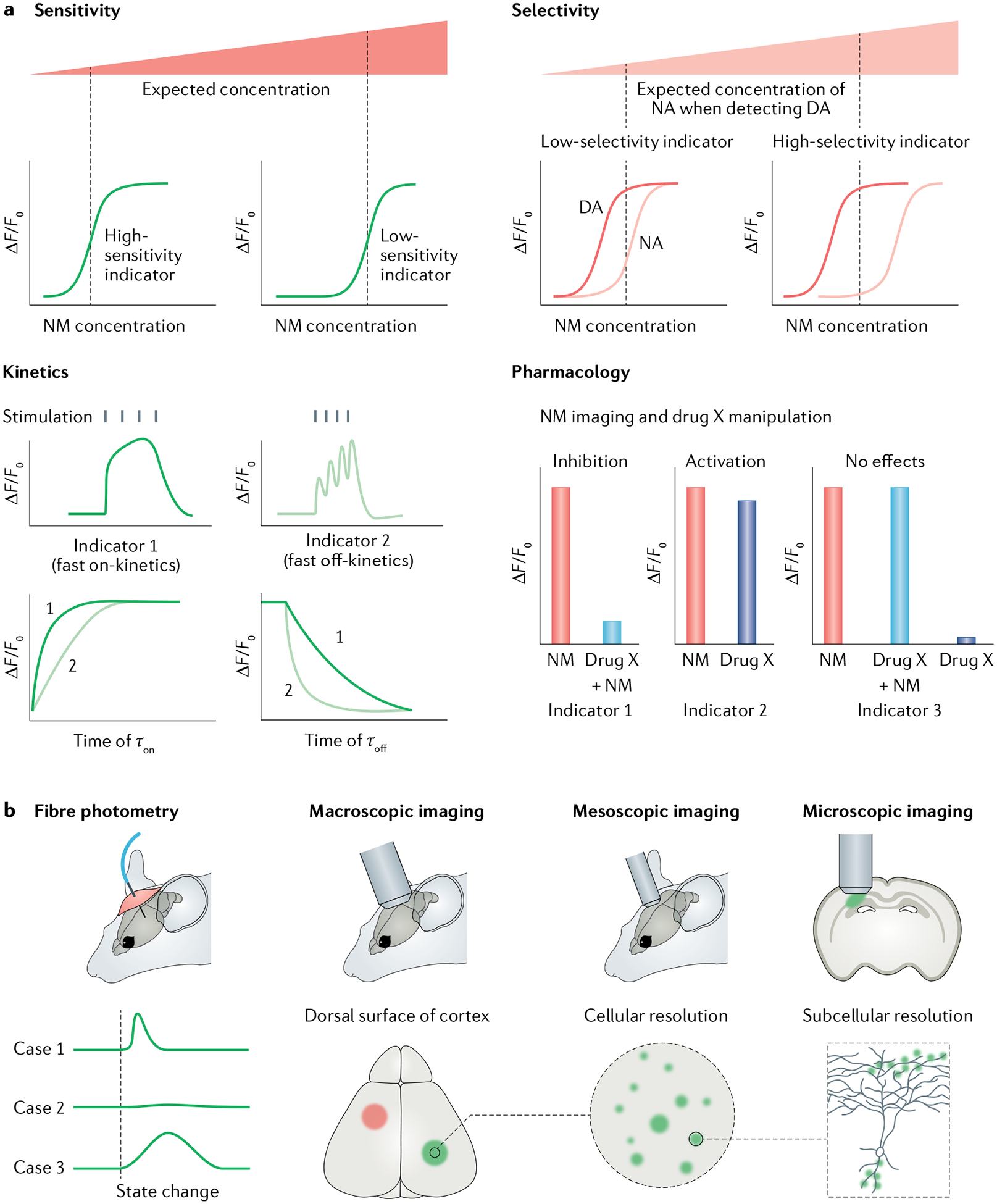
a | Parameters for choosing a genetically encoded neurotransmitter or neuromodulator (NT/NM) indicator (GENI). First, is the GENI sensitive enough for measuring NTs or NMs of interest? Generally, if two sensors showed comparable maximum responses (ΔF/F0), the sensor with higher affinity should be used to detect NT or NM at low concentration. Second, is the sensor selective enough? For example, if users want to detect dopamine (DA) levels, a DA sensor that could better separate DA and noradrenaline (NA) should be chosen. Third, is the sensor fast enough to capture NT or NM dynamics? GENIs with fast on-kinetics (τon) will have relatively large sensor responses to brief release events. GENIs with fast off-kinetics (τoff) are preferred to track the dynamics of NTs and NMs in response to closely related events or rapid changes in behaviour. Fourth, is the GENI compatible with any pharmacological manipulations used? GENIs, especially G protein-coupled receptor (GPCR)-based indicators, are engineered using NT or NM receptors and thus inherit their parental GPCRs’ pharmacological properties. Therefore, GPCR-based indicators are incompatible with certain pharmacological manipulations. b | Imaging modalities and data interpretations. Fibre photometry recording is the method of choice for detecting GENI signals in the brain region of interest, in freely moving animals. When used with GENIs with fast kinetics, fibre photometry recording can report NT or NM dynamics during animal state changes (such as during sleep–wake transitions and learning). Fibre photometry measures bulk fluorescence emitted from a radius of several hundred microns and therefore has relatively poor spatial resolution. One advantage of GENIs is that they can be imaged at multiple spatial scales, ranging from the whole brain to subcellular levels, when combined with different imaging modalities.
