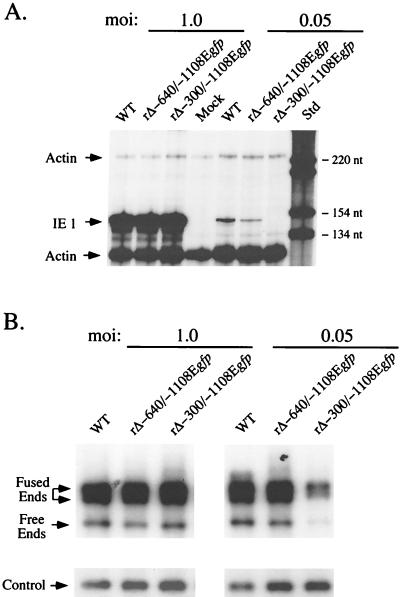
FIG. 7.
Analysis of productivity of IE1 RNA and viral DNA by WT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp in HFF cells at high or low MOI. (A) Analysis of viral IE1 RNAs. HFF cells were infected in parallel with WT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp at MOIs of 1.0 and 0.05. Total cellular RNA was harvested at 8 hpi. Equal amounts of RNAs (20 μg) were subjected to RNP assay, using both IE1- and actin-specific riboprobes, as described previously (17, 31, 56) (see Materials and Methods). RNA of uninfected HFF cells (Mock) served as a control. Positions of protected actin (230 and 122 nt) and IE1 (145 nt) RNAs (denoted by arrows), as well as selected size markers (Std), are shown. (B) Analysis of viral DNAs. HFF cells were infected with WT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp at MOIs of 1.0 and 0.05. Infections were performed in parallel with those shown in panel A. Infected cell DNA was isolated at 4 dpi, digested with HindIII, and subjected to Southern blotting using the 32P-labeled T or λ probe described in the legend to Fig. 4A. λ DNA served as an internal control as detailed for Fig. 4A. Arrows indicate positions of 9.7-kb RL free ends, 17.2- and 13-kb RL fused ends, and internal λ control. At an MOI of 0.05, abundance of RL fused and free ends of rΔ-300/-1108Egfp differed by approximately 3- and 2.7-fold, respectively, from those of WT or rΔ-640/-1108Egfp.