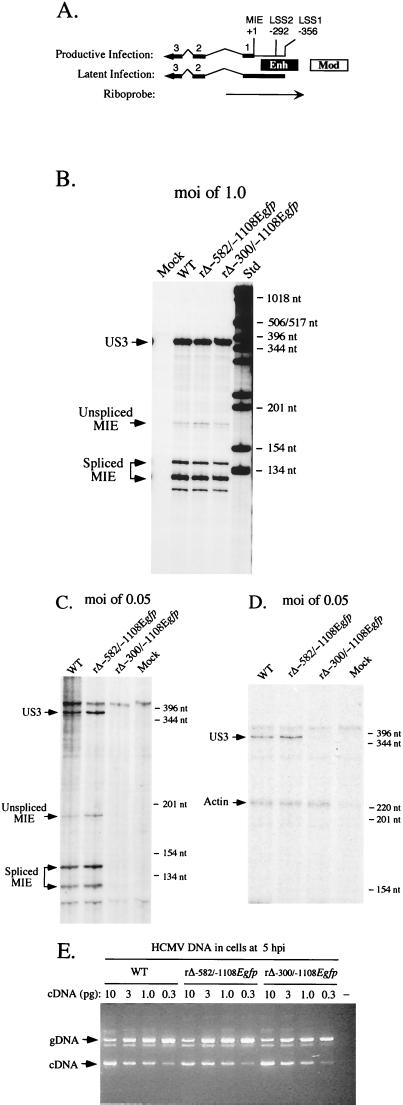
FIG. 8.
Analysis of both MIE and US3 RNAs produced by WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp in HFF cells at high or low MOI. (A) Schematic diagram of MIE riboprobe. Position of MIE riboprobe (rightward arrow) in relation to +1 start site of MIE RNA is depicted. LS RNAs arise from two sites, LSS1 and LSS2 at base positions -356 and -292, respectively (21, 22). Both MIE and LS RNAs (leftward arrows) possess exons (thick lines; only exons 1, 2, 3, and L [UL126 ORF] are depicted) and introns (thin lines). The riboprobe extends from +171 to −582, spanning the enhancer (Enh), exon 1, and part of intron 1. It is predicted to protect unspliced and spliced MIE RNAs of 170 and 120 nt, respectively. It also can protect an alternatively spliced MIE RNA of ∼140 that was noted previously by Stenberg et al. (49). Mod, modulator. (B) Analysis of MIE and US3 RNAs in infected HFF cells at an MOI of 1.0. Cells were infected in parallel by WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp. Total cellular RNA (20 μg) was isolated at 8 hpi and subjected to RNP assay, using both MIE- and US3-specific riboprobes (see Materials and Methods). RNA of uninfected HFF cells (Mock) served as a control. Positions of protected US3 (370 nt) and unspliced (170) and spliced (120 and 140 nt) MIE RNAs (denoted by arrows), as well as selected size markers (Std), are shown. (C) Analysis of MIE and US3 RNAs in infected HFF cells at an MOI of 0.05. Cells were infected in parallel by WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp. Total cellular RNA (25 μg) was isolated at 8 hpi and subjected to RNP assay, using both MIE- and US3-specific riboprobes. (D) Analysis of US3 and actin RNAs in infected HFF cells at an MOI of 0.05. Total cellular RNA (25 μg) isolated from WT-, rΔ-582/-1108Egfp-, and rΔ-300/-1108Egfp-infected HFF cells and employed in studies shown in panel C was analyzed by RNP assay, using US3- and cellular actin-specific riboprobes. Positions of protected US3 (370 nt) and actin (230 nt) RNAs (denoted by arrows), as well as selected size markers, are shown. (E) Viral entry into HFF cells. HFF cells were infected at an MOI of 0.05 with WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp in parallel to those shown in panels C and D. Infected cells were thoroughly washed, and DNA was isolated at 5 hpi. This cell-associated DNA was then subjected to QC-PCR that amplifies a region spanning intron 3 of the HCMV IE1 gene (see Materials and Methods). Amplification of viral genomes (gDNA) and IE1 cDNA generates 387- and 217-bp products, respectively. Each QC-PCR mixture contained 600 ng of cell-associated DNA and the indicated amount (10, 3.0, 1.0, or 0.3 pg) of the 1,765-bp IE1 cDNA. QC-PCR products were fractionated on an agarose gel containing ethidium bromide. All steps were done in parallel. The negative control (−) contains uninfected cell DNA but no IE1 cDNA. Arrows indicate positions of 387 bp (gDNA) and 217 bp (cDNA) PCR products, as gauged by DNA size markers that are not shown.