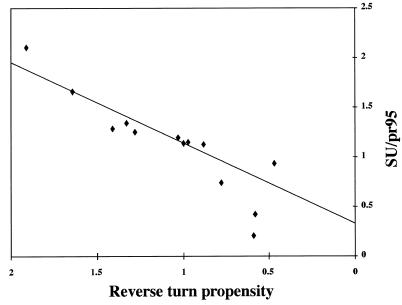
FIG. 4.
Processing of mutant EnvAs into SU (gp85) and TM (gp37) subunits. Cells expressing wild-type and mutant EnvAs were radiolabeled with Tran35S-label, lysed, subjected to immunoprecipitation using the anti-EnvA tail antibody, and treated with N-glycosidase F. Proteins were resolved by SDS-PAGE, visualized using a PhosphorImager, and quantified using ImageQuant. The data were subjected to linear regression analysis and fit best to the linear equation y = 0.81× + 0.33 with r2 = 0.84. The data (triangles) represent the ratio of gp85 to unprocessed pr95. The solid line represents the linear least squares fit of the data. The two points significantly below the line are for the poorly trimerized F and L mutants.