Extended Data Fig. 9 |. Integrated model for how spliceosome activity and proximity to nuclear speckles impact kinetics of splicing.
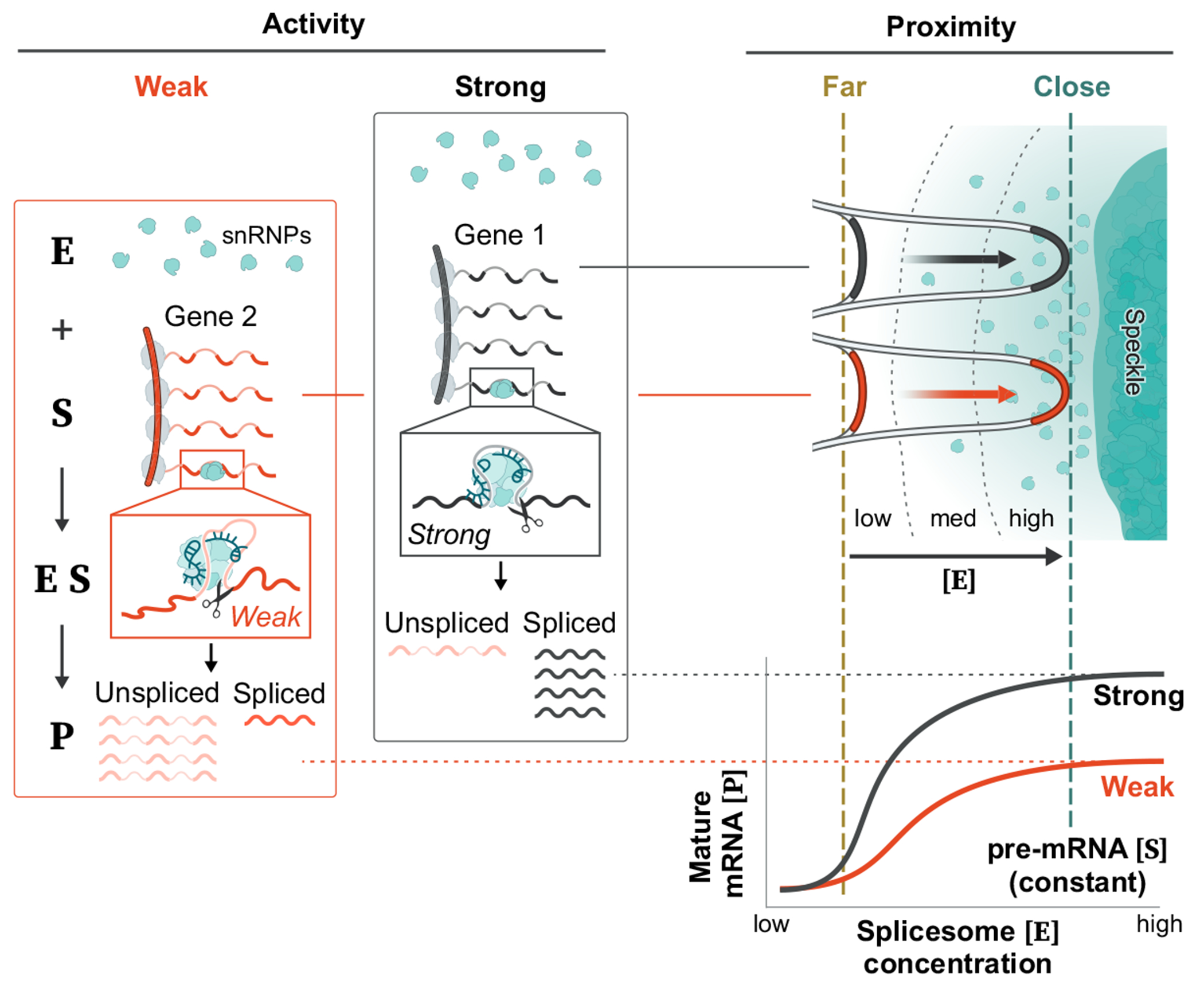
There are two components impacting the kinetics of splicing – spliceosome concentration and spliceosome activity. (i) Proximity to nuclear speckles impacts the concentration of spliceosomes at a given pre-mRNA, such that genes that are close to speckles will have higher spliceosome concentration than genes that are far from speckles. (ii) In contrast, splice site strength is defined by the activity of the spliceosome at the splice site53. In this way, spliceosomes engaged at ‘strong’ splice sites would have higher activity while ‘weak’ splice sites would have lower activity. These two components would be expected to have different effects on the kinetics of splicing. Specifically, modulating activity (splice site strength) would be expected to impact the maximum output of the reaction. Conversely, modulating concentration (speckle proximity) would be expected to impact the efficiency of each reaction.
