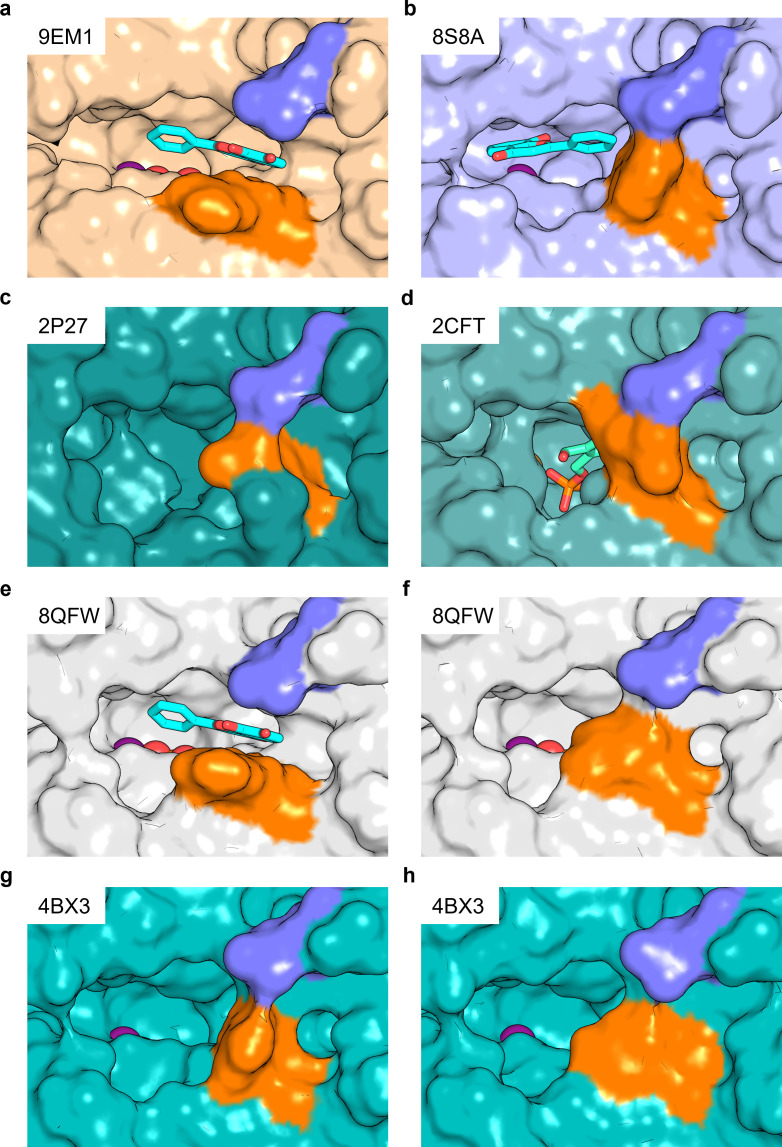
Figure 3. X-ray crystal structures of human pyridoxal 5’-phosphate phosphatase (PDXP) in complex with 7,8-dihydroxyflavone (7,8-DHF).
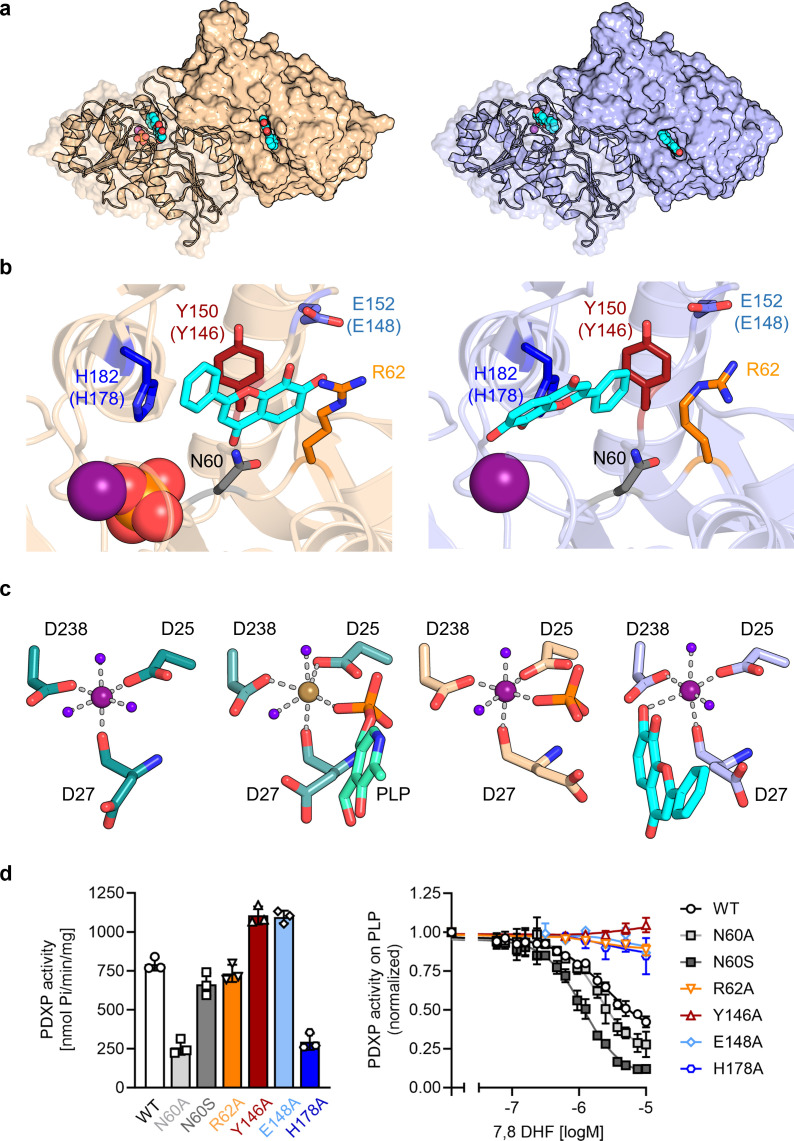
(a) The models were refined to a resolution of 1.5 Å for full-length human 7,8-DHF-PDXP with phosphate (PDB code 9EM1, colored in wheat yellow, left panel) and 1.5 Å for full-length human 7,8-DHF-PDXP without phosphate (PDB code 8S8A, colored in light blue, right panel). One protomer of each homodimeric PDXP is shown in cartoon representation and the other protomer in surface representation. 7,8-DHF is displayed in sphere representation with its C-atoms in cyan. Mg2+ ions are shown as deep purple spheres and phosphate ions are shown in sphere representation with the phosphorous atom in orange. (b) Orientation of 7,8-DHF in the active sites of human 7,8-DHF-PDXP in the presence or absence of phosphate. Structural details of bound 7,8-DHF and adjacent residues of the active sites are shown. Left, phosphate-containing 7,8-DHF-PDXP (wheat yellow, cartoon representation). Right, phosphate-free 7,8-DHF-PDXP (light blue, cartoon representation). 7,8-DHF is shown in stick representation (cyan C-atoms). The corresponding amino acids in murine PDXP are given in parentheses (see also Figure 3—figure supplement 1e and f). (c) Comparison of the Mg2+ coordination spheres. From left to right: human apo-PDXP (PDB: 2P27), human PDXP in complex with pyridoxal 5’-phosphate (PLP) (PDB: 2CFT), human PDXP in complex with 7,8-DHF in the presence of phosphate (PDB: 9EM1), human PDXP in complex with 7,8-DHF in the absence of phosphate (PDB: 8S8A). The catalytically essential Mg2+ is shown as a deep purple sphere. In 2CFT, Mg2+ was exchanged for Ca2+, which is shown here as a light brown-colored sphere. Water molecules are shown as blue spheres. (d) Verification of 7,8-DHF-PDXP interactions. Left panel, phosphatase activity of purified PDXP or the indicated PDXP variants. Data are mean values ± SD of n=3 biologically independent experiments. Right panel, determination of the IC50 values of 7,8-DHF for purified PDXP or the indicated PDXP variants. Data are mean values ± SD of n=3 biologically independent experiments. Apparently missing error bars are hidden by the symbols.
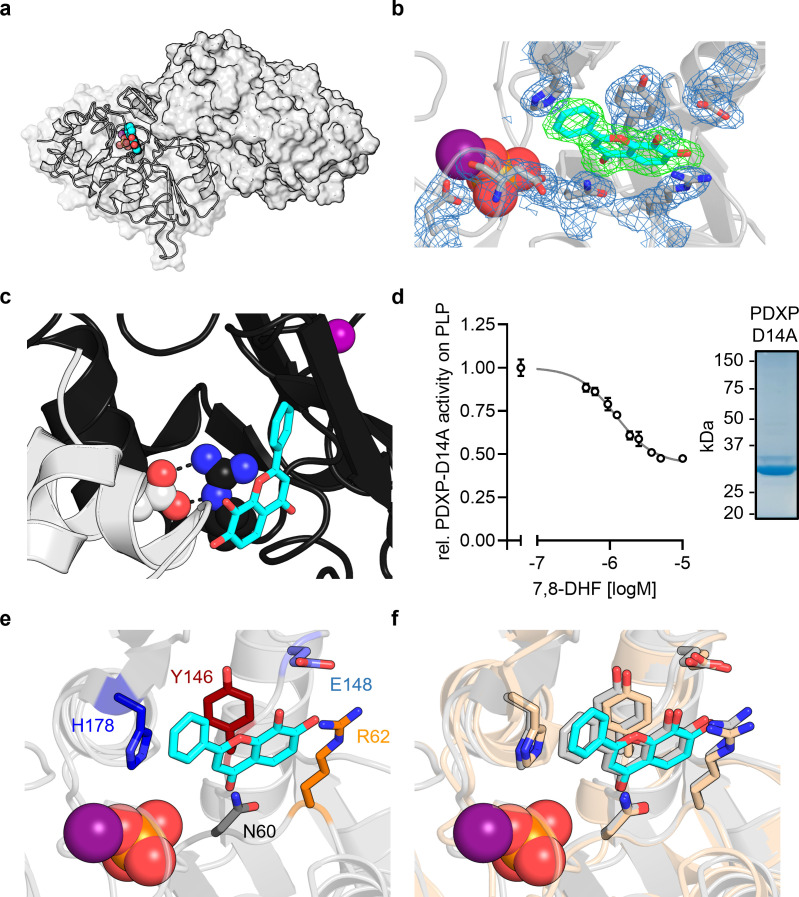
Figure 3—figure supplement 1. X-ray crystal structures of murine pyridoxal 5’-phosphate phosphatase (PDXP) in complex with 7,8-dihydroxyflavone (7,8-DHF).
Figure 3—figure supplement 2. 7,8-Dihydroxyflavone (7,8-DHF) coordination in pyridoxal 5’-phosphate phosphatase (PDXP).
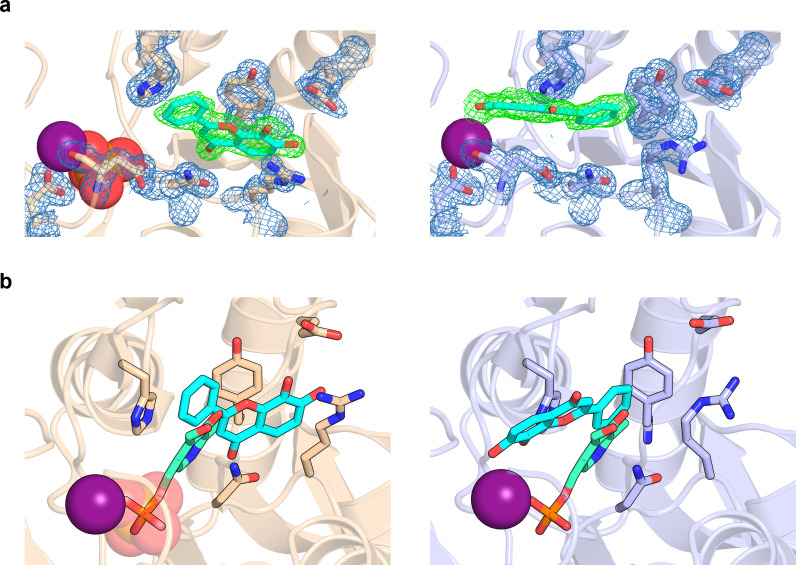
Figure 3—figure supplement 3. Alignment of human and murine pyridoxal 5’-phosphate phosphatase (PDXP).

Figure 3—figure supplement 4. Salt bridge formation between Glu152 (Glu148) and Arg62 gates the active site entrance in pyridoxal 5’-phosphate phosphatase (PDXP).
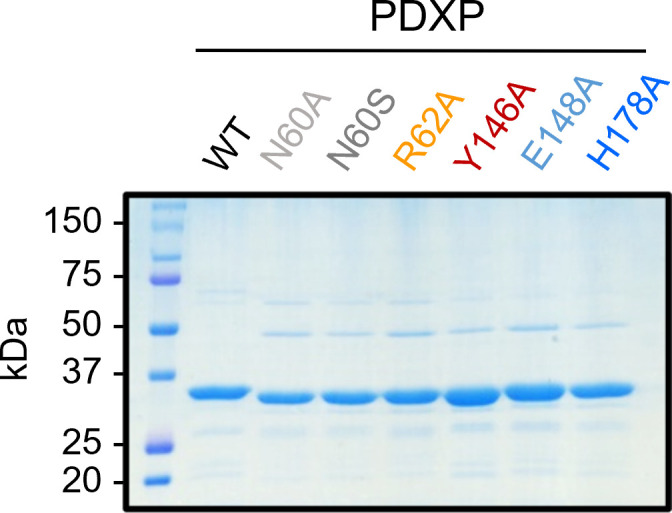
Figure 3—figure supplement 5. Purity of the employed pyridoxal 5’-phosphate phosphatase (PDXP) and PDXP variants.