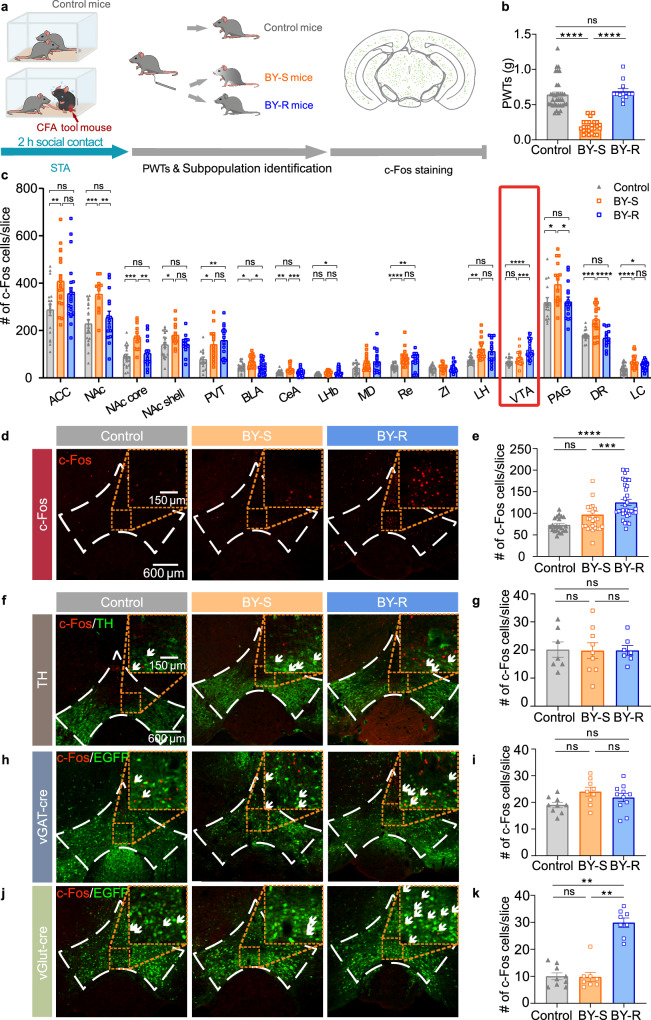
Fig. 2. Activation of VTA glutamatergic neurons in the resilient mice.
a Experimental timeline (adapted from The Mouse Brain in Stereotaxic Coordinates by Paxinos and Franklin). b PWTs (n = 35, 26 and 12 mice). Control versus BY-S, P = 8.2 × 10−10; BY-S versus BY-R, P = 4.9 × 10−8. c Quantification of c-Fos protein expression in different brain regions (n = 15–35 slices from 5–9 mice). d, e Representative immunofluorescent images and quantitative data of VTA c-Fos protein expression (n = 22, 25 and 33 slices from 7, 8 and 9 mice); Control versus BY-R, P = 2.5 × 10−7; BY-S versus BY-R, P = 0.0007. f, g Representative immunofluorescent images and quantitative data of c-Fos protein expression in VTA dopaminergic neurons (n = 9, 8 and 8 slices from 3, 3 and 3 mice); Control versus BY-R, P = 0.9998; BY-S versus BY-R, P > 0.9999. h, i Representative immunofluorescent images and quantitative data of c-Fos protein expression in VTA GABAergic neurons (n = 7, 9 and 7 slices from 3, 3 and 3 mice). Control versus BY-R, P = 0.3702; BY-S versus BY-R, P = 0.5458. j, k Representative immunofluorescent images and quantitative data of c-Fos protein expression in VTA glutamatergic neurons (n = 9, 9 and 11 slices from 3, 3 and 3 mice). Control versus BY-R, P = 0.0017; BY-S versus BY-R, P = 0.0015. White arrows indicate overlapped neurons. The staining was repeated twice with similar results. Scale bar: 600 and 150 µm. ACC anterior cingulate cortex; NAc nucleus accumbens; PVT paraventricular thalamic nucleus; BLA basolateral amygdala; CeA central amygdala; LHb lateral habenula; MD mediodorsal thalamic nucleus; Re reuniens thalamic nucleus; ZI zona incerta; LH lateral hypothalamus; VTA ventral tegmental area; PAG periaqueductal gray; DR dorsal raphe nucleus; LC locus coeruleus; TH tyrosine hydroxylase. Data are presented as the mean ± s.e.m. Data analyzed by (b, e, g) Kruskal–Wallis test with Dunn’s multiple comparisons test, or (i, k) one-way ANOVA with Tukey’s multiple comparisons test. ns no significance, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Statistical details are presented in Supplementary Table 5. Source data are provided as a Source Data file.