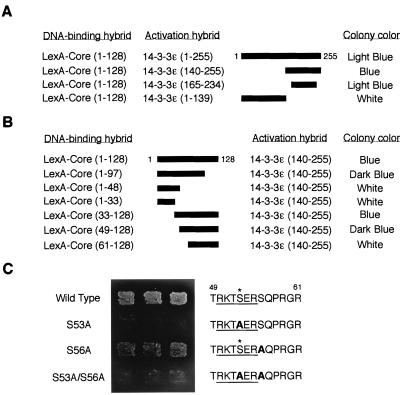
FIG. 1.
Interaction of HCV core protein with 14-3-3 protein in the yeast two-hybrid system. (A) Determination of binding domain within 14-3-3 protein. Various activation hybrid mutants were expressed in the yeast strain EGY48 with LexA-Core protein (amino acids 1 to 128). A schematic of these 14-3-3 deletion mutants is shown with thick bars. Protein interaction was monitored by activation of the lacZ reporter gene (determined by colony color; blue colony represents a positive interaction) on an SC plate containing 2% galactose and 80 mg of X-Gal per liter with 3 days of incubation at 30°C. (B) Determination of binding domain within HCV core protein. LexA fusion proteins containing the indicated sequences of HCV core protein were expressed with an activation domain hybrid of 14-3-3 protein, 14-3-3ɛ (amino acids 140 to 255), in the yeast strain EGY48. Interactions were monitored as described for panel A. (C) HCV core protein interacts with 14-3-3 protein in a phosphoserine-dependent manner in yeast. LexA fusion proteins containing wild-type and S53A, S56A, and S53A/S56A mutants of HCV core protein (amino acids 1 to 128) were expressed with 14-3-3ɛ, an activation hybrid of 14-3-3 protein (amino acids 140 to 255), in the yeast strain EGY48, and the phosphoserine-dependent interaction was monitored by activation of leucine reporter gene and by colony color on a leucine-deficient SC plate containing galactose and X-Gal for 3 days. The putative 14-3-3 binding motifs (underlined) within HCV core proteins are indicated on the right. Substituted alanine residues are indicated by boldface type. Asterisks indicate phosphorylation sites for PKA and PKC.