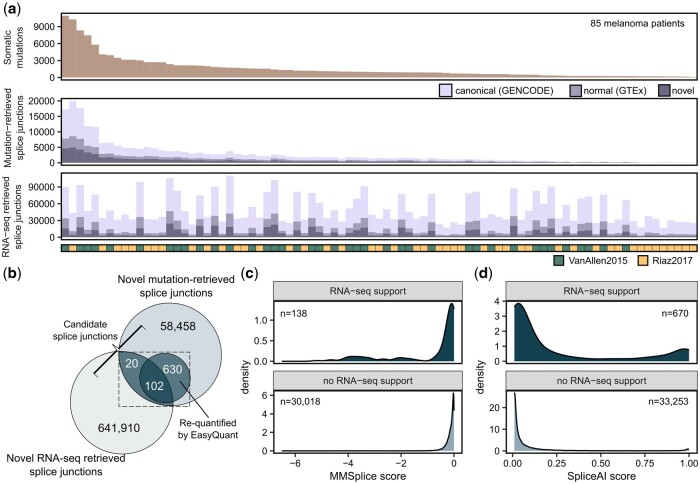
Figure 2.
Identification of mutation-retrieved splice junctions supported by RNA-seq in melanoma samples. (a) The number of somatic mutations, mutation-retrieved splice junctions and RNA-seq splice derived junctions per sample in the discovery cohort of 85 melanoma samples. (b) Overlap of mutation-retrieved splice junctions with those that were found in RNA-seq by SplAdder or LeafCutter and those that were requantified in RNA-seq with EasyQuant. Mutation-retrieved splice junctions with RNA-seq support (i.e. by SplAdder, LeafCutter, or EasyQuant) were defined as candidate splice junctions. (c, d) The distribution of (c) MMSplice and (d) SpliceAI scores of candidate splice junctions with RNA-seq support and mutation-retrieved splice junctions without RNA-seq support.