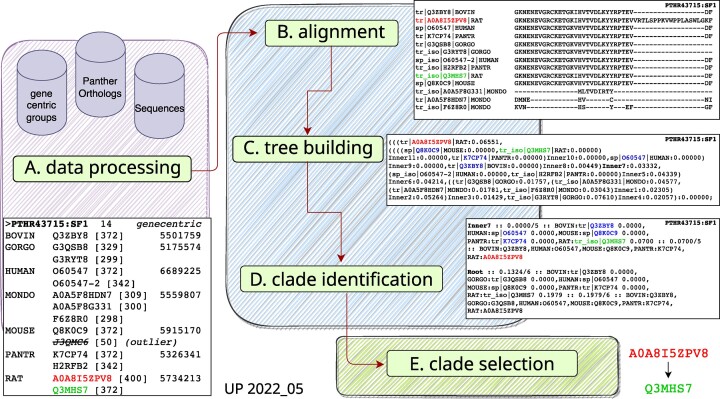
Figure 2.
The ortho2tree analysis pipeline. (A) Gene-Centric groups for Panther orthologs (22) are used to extract orthologous canonical and isoform sequences. Panther Ortholog IDs are used to group genes among organisms; Gene-Centric groups are used to identify canonical and isoform sequences associated with each Panther Ortholog set. (B) Orthologous canonical proteins and orthologs are aligned using muscle (23). (C) A gap-distance tree (see Materials and Methods) is constructed using the BioPython3.8 (24) functions DistanceCalculator, DistanceMatrix and DistanceTreeConstructor. (D) A heuristic clade search script examines the gap-distance tree and identifies low-cost clades with sequences from different proteomes. (E) The list of low-cost clades is scored based on the number of proteomes, the cost of the clade, the number of UniProtKB/Swiss-Prot entries from human or mouse, and length consistency. The highest scoring clades from each distinct gene set is then plotted on a tree (Figure 3).