Fig. 5 |. Diversity of wyrdviruses.
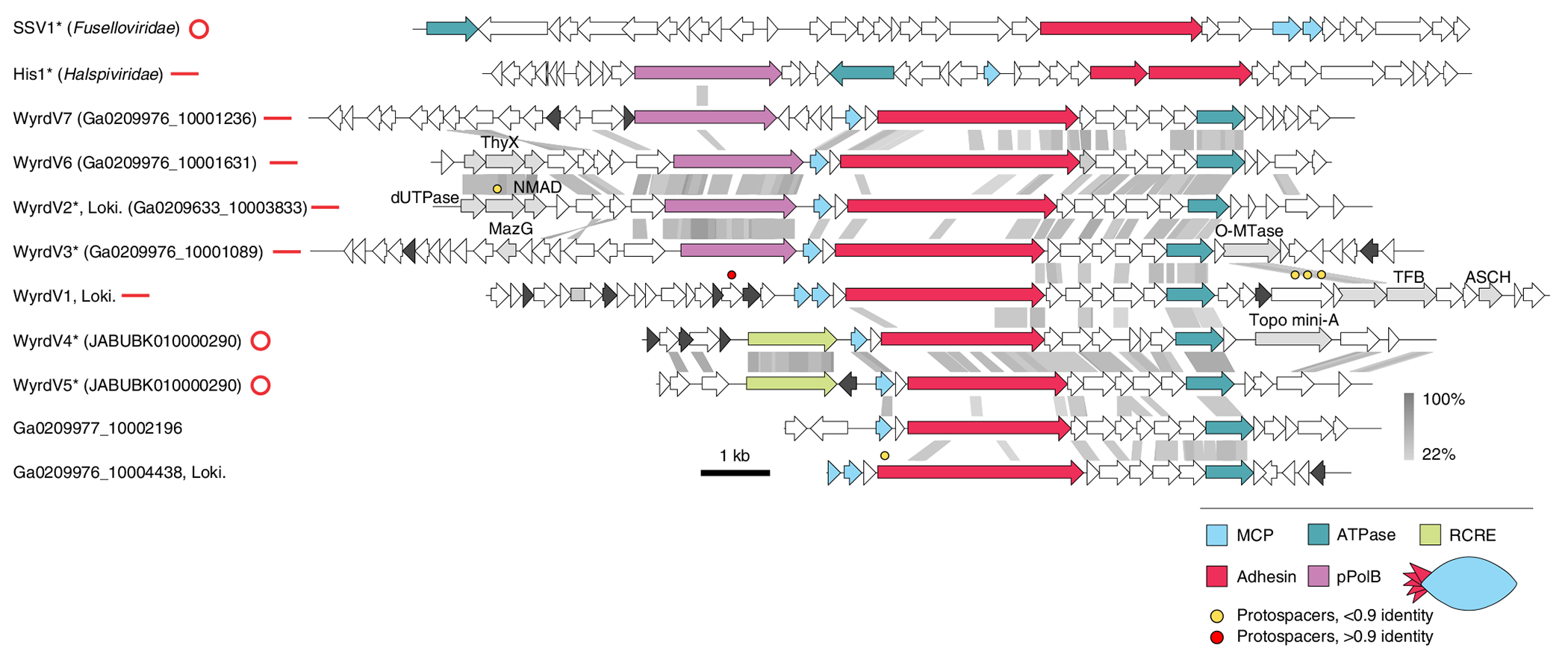
Genome maps of wyrdviruses, fusellovirus SSV1 and halspivirus His1. Homologous genes are shown using the same colours, with the key provided at the bottom. Also shown is the deduced schematic organization of the skuldvirus virion with colours marching those of the genes encoding the corresponding proteins. Coloured circles indicate the positions of protospacers. Genes encoding putative DNA-binding proteins with Zn-binding and HTH domains are coloured black and grey, respectively. Grey shading connects genes displaying sequence similarity at the protein level, with the percentage of sequence identity indicated by different shades of grey (see scale on the right). Asterisks denote complete genomes assembled as circular contigs or linear contigs with terminal inverted repeats. The topology of each genome is shown next to the corresponding name: circles for circular genomes, lines for linear genomes. ThyX, thymidylate synthase X; NMAD, nucleotide modification-associated domain 1 protein; O-MTase, carbohydrate-specific methyltransferase; TFB, transcription initiation factor B; ASCH, ASC-1 homology domain; pPolB, protein-primed family B DNA polymerase. Genome maps of SSV1 and His1 viruses are shown for comparison.
