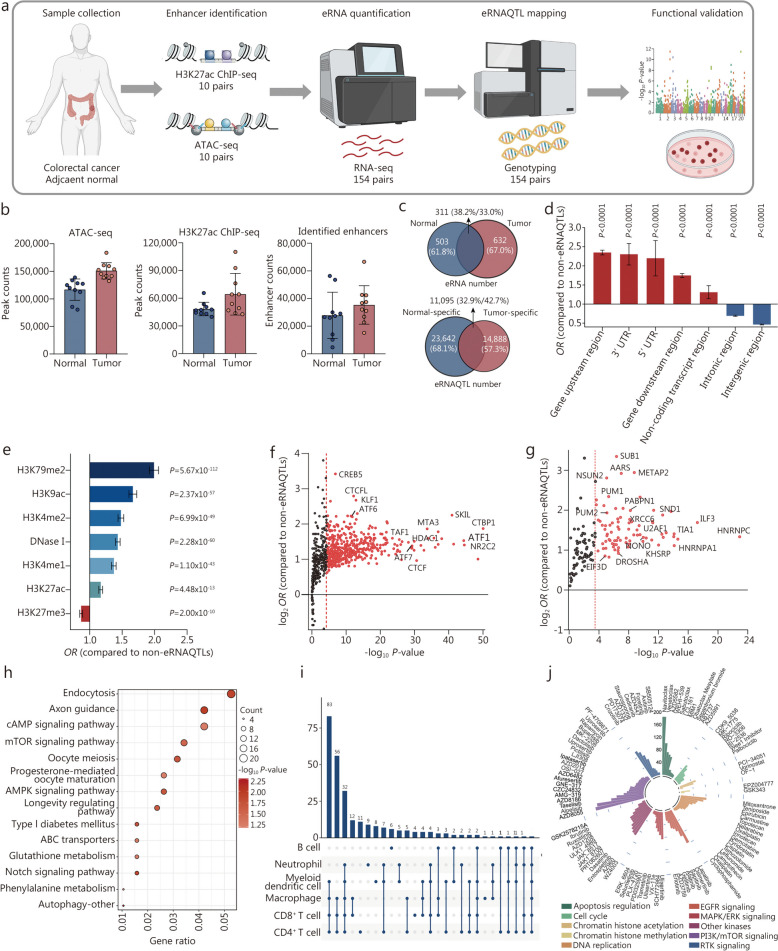
Fig. 3.
Validation of eRNAQTL profiles in Chinese colorectal cancer (CRC) samples. a Flowchart of enhancer identification and eRNAQTLs mapping in our CRC discovery set. ATAC-seq and H3K27ac ChIP-seq signals of 10 pairs of CRC tumor and adjacent normal tissues were integrated to identify active enhancer elements. Then, RNA-seq data of 154 pairs of tumor and normal tissues were mapped to these enhancer regions for quantifying eRNA transcription. By combining genotype data, the first Chinese eRNAQTL profiles were established in CRC. b The number of ATAC-seq peaks, H3K27ac ChIP-seq peaks, and active enhancers in 10 pairs of tumor and adjacent normal tissues. c Venn diagram showing the number of eRNAs (top) and eRNAQTLs (bottom) identified in 154 pairs of normal and tumor tissues. d Enrichment analysis of tumor eRNAQTL SNPs in different genomic categories. P-values were calculated by a two-tailed Fisher’s exact test. Bars indicate 95%CI. e Enrichment analysis of tumor eRNAQTLs among variants within regulatory elements. P-values were calculated by a two-tailed Fisher’s exact test. Bars indicate 95%CI. f Plots of -log10 P-value (X-axis) and -log2 OR (Y-axis) were obtained from enrichment analysis of tumor eRNAQTLs among variants within TF binding sites. The red dashed line indicates P = 0.05/801 (6.24 × 10−5) (Bonferroni-corrected P-value threshold, binding sites for a total of 801 TFs were tested). g Enrichment of eRNAQTL SNPs at RBP binding sites by two-tailed Fisher’s exact test. The red dashed line indicates the Bonferroni-corrected P-value threshold according to the number of RBPs tested (150 RBPs). h KEGG pathway analysis for target genes of eRNAQTL-eRNAs in CRC. The circle size represents the number of genes enriched in each pathway, and the color indicates the -log10 P-value of enrichment. i Number of eRNAQTL-eRNAs associated with immune cell fractions, calculated by partial correlation coefficient with tumor purity adjusted. j Circle plot summarized the number of eRNAQTL-eRNAs associated with pharmaceutical targets in different categories. The count numbers are displayed with different colors by the cancer signaling pathway. ATAC-seq assay for transposase-accessible chromatin with high throughput sequencing, ChIP-seq chromatin immunoprecipitation sequencing, CI confidence interval, RBP RNA binding protein, KEGG Kyoto Encyclopedia of Genes and Genomes, OR odds ratio, CRC colorectal cancer, SNP single nucleotide polymorphism, eRNAQTL eRNA quantitative trait locus, MAPK/ERK mitogen-activated protein kinase/extracellular signal-regulated kinase, EGFR epidermal growth factor receptor, RTK receptor tyrosine kinase, PI3K/mTOR phosphoinositide 3-kinase/mammalian target of rapamycin pathway, cAMP cathelicidin antimicrobial peptide, ABC transporters ATP-binding cassette transporters, AMPK adenosine 5’-monophosphate (AMP)-activated protein kinase, mTOR mammalian target of rapamycin