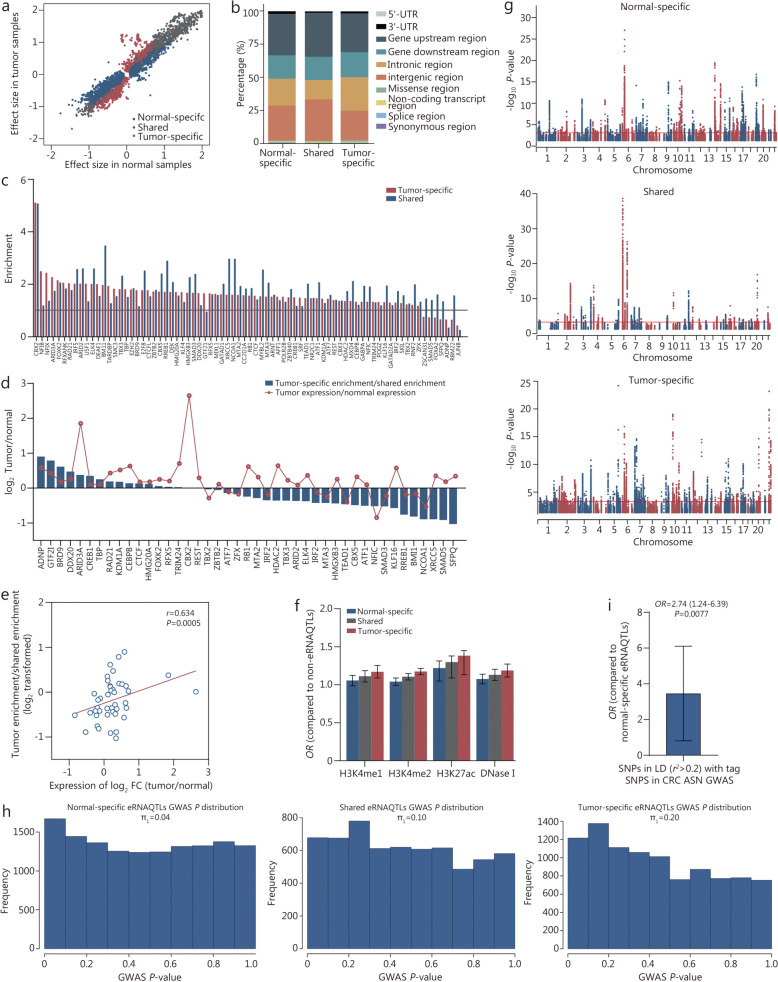
Fig. 4.
Comparison between eRNAQTLs in normal and tumor-derived colorectal tissues. a Effect sizes of eRNAQTLs identified in normal samples vs. tumor samples. b The stacked bar charts indicate the percentage of 10 functional categories for each type of eRNAQTLs. c The enrichment of tumor-specific eRNAQTLs and shared eRNAQTLs in TF binding sites. The bar plot is ordered by the tumor-specific enrichment. The null (frequency and variant type matched) is represented as the black horizontal line. d The ratio of the tumor-specific enrichment to the shared enrichment (in log scale) for significantly differentially expressed transcription factors (TFs) are shown as blue bars. The red line depicts the differential expression fold change of the TFs. e Correlation between the enrichment in TF binding sites and fold change (FC) of differentially expressed TFs. f Enrichment analysis of eRNAQTLs among variants within 4 epigenomic marks, including H3K4me1, H3K4me2, H3K27ac, and DNase I hypersensitive sites. P-values were calculated by a two-tailed Fisher’s exact test. g Manhattan plot of -log10 P-value from the results of three types of eRNAQTLs in our CRC samples (normal-specific eRNAQTLs, shared eRNAQTLs, and tumor-specific eRNAQTLs). The eRNAQTL P-values (-log10) of the SNPs (Y-axis) are presented according to their chromosomal positions (X-axis). h CRC GWAS P-value distributions and statistics for normal-specific, shared, and tumor-specific eRNAQTLs. i Enrichment analysis of tumor-specific eRNAQTL SNPs among CRC risk loci in Asian populations. The P-value was calculated by two-tailed Fisher’s exact tests. OR is depicted on the Y-axis and bars indicate 95%CIs. OR odds ratio, CRC colorectal cancer, CI confidence interval, ASN Asian, eRNAQTL eRNA quantitative trait locus, SNP single nucleotide polymorphism, GWAS genome-wide association study