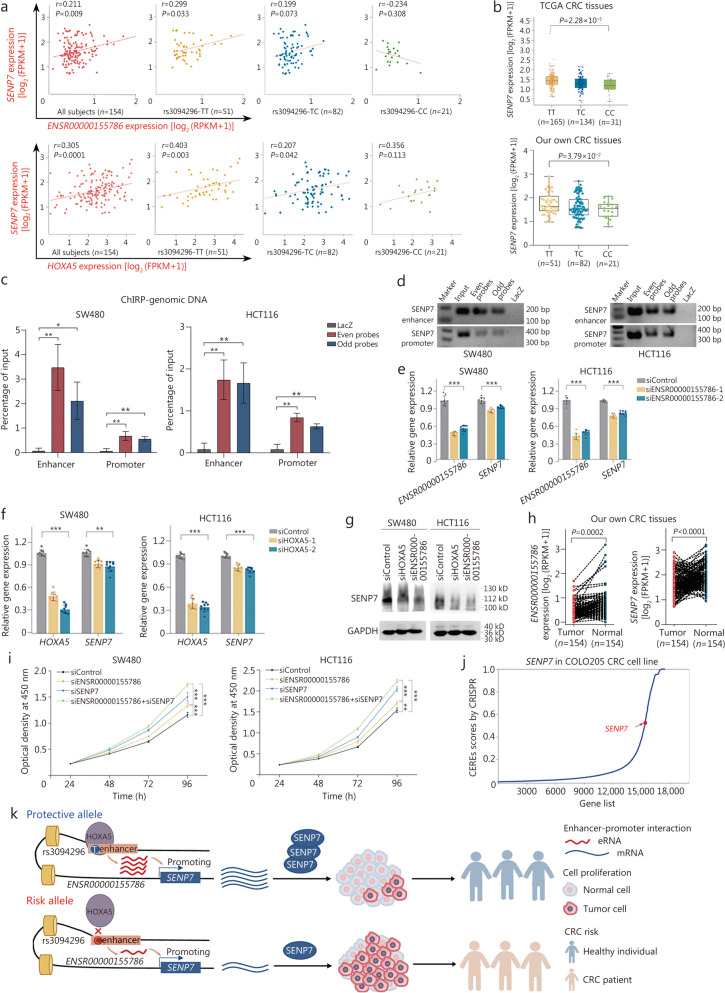
Fig. 6.
eRNA ENSR00000155786 inhibits colorectal cancer (CRC) cell proliferation by activating SENP7 expression. a Scatter plots show the correlations between ENSR00000155786 and SENP7 expression, as well as the correlations between HOXA5 and SENP7 expression in our CRC tumor tissues both of them, are stratified by SNP rs3094296 genotype. All P-values and correlation coefficients were calculated by Spearman’s correlation analysis. b eQTL analyses demonstrate the correlation between the rs3094296 genotype and the expression of SENP7 in the TCGA CRC samples and our own CRC tissues. Data were shown as the median (minimum to maximum). ChIRP assay was performed using two different sets of antisense probes (“even group” and “odd group”) that detected ENSR00000155786 or control probes (anti-LacZ) in CRC cell lines. The enrichment of genomic DNA in the ChIRP and input samples was measured by qPCR (c) and agarose gel electrophoresis (d). Data were shown as the mean ± SD. P-values were calculated by a two-sided Student’s t-test. The histogram displays the effect of ENSR00000155786 knockdown on the expression of SENP7 (e) and the effect of HOXA5 knockdown on the expression of SENP7 (f) in SW480 and HCT116 cells. Data were presented as the mean ± SD from three repeated experiments with three technical replicates. All P-values were calculated using a two-sided Student’s t-test. g Western blotting analysis shows that the knockdown of HOXA5 and ENSR00000155786 reduced the protein expression level of SENP7 in SW480 and HCT116 cells. h ENSR00000155786 and SENP7 are significantly decreased in tumor tissues compared with normal tissues from our own CRC tissues. P-values were calculated by a paired two-sided Student’s t-test. i Cell proliferation assay with knockdown of ENSR00000155786 and SENP7 in SW480 and HCT116 cells. Results were shown as the means ± SEM from three experiments, each with six replicates. P-values were calculated from a two-sided Student’s t-test by comparing with controls in 96 h. j SENP7 is essential for cell growth with higher CERE scores in COLO205 CRC cell lines from the data of genome-wide CRISPR/Cas9-based loss-of-function screen. Higher CERE scores demonstrate an elevated dependency of cell viability on given genes. k The graph shows the mechanism that eRNAQTL rs3094296 contributes to a decreased risk of CRC by playing allele-specific effect to transcribe eRNA ENSR00000155786, which further exerts a transcriptional activator promoting target gene SENP7 expression, then these two genes display a synergistic effect to inhibit tumor cells proliferation. *P < 0.05, **P < 0.01, ***P < 0.001. FPKM fragments per kilobase of exon model per million mapped fragments, RPKM reads per kilobase per million mapped reads, HOXA5 homeobox A5, SENP7 sentrin-specific protease 7, ChIRP chromatin isolation by RNA purification, CERES CRISPR enrichment-based screening, SEM standard error of the mean