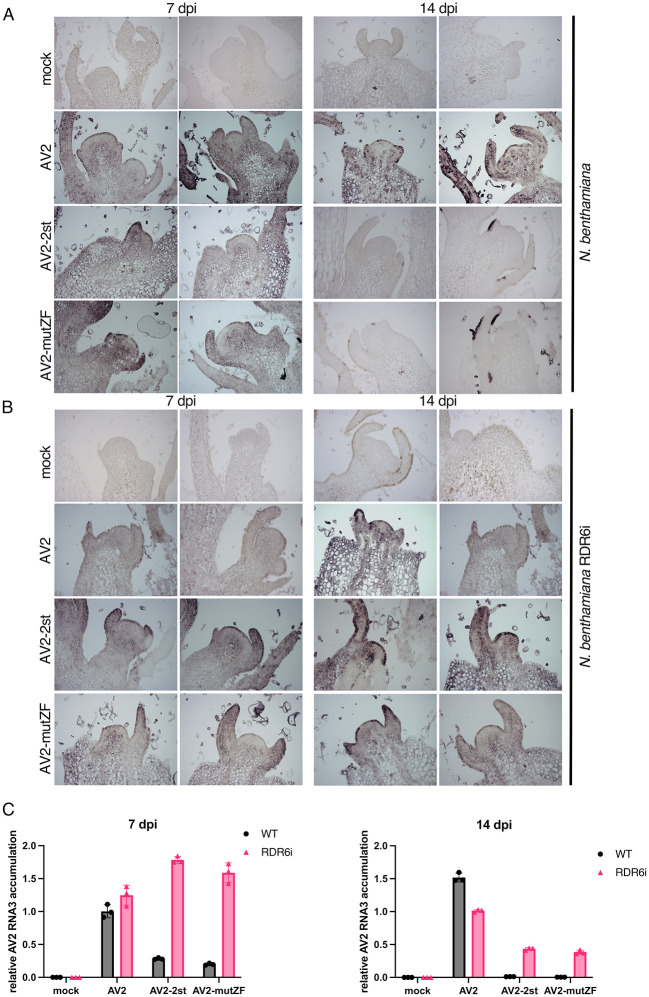
Fig 9. Detection of RNA3 in longitudinal sections of the top-most shoot apices.
(A) Wild type N. benthamiana plants were infected with AV2, AV2-2st or AV2-mutZF, and samples were collected at 7 and 14 dpi. AV2 RNA was detected by in situ hybridisation using the digoxigenin (Roche Diagnostics GmbH) labelled in vitro transcribed RNA fragment complementary to the AV2 CP ORF. Darker blue-purple areas indicate hybridisation signal indicative of viral RNA. (B) As for panel A but using RDR6i transgenic N. benthamiana plants. In A and B two representative sections are shown for each treatment at each time-point. (C) RT-qPCR analysis of RNA3 accumulation in apical tissues of infected plants. Samples were collected at the same time points as for in situ hybridisation. Shoot apices of five infected plants were pooled together for RNA extraction. Nb ACT-b (GI:380505031) was used as a reference gene. Values are expressed in arbitrary units and represent the mean +/− SEM of three technical replicates.