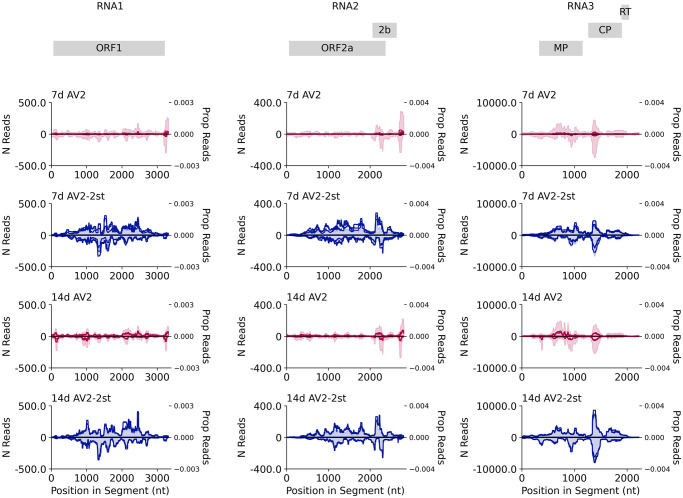
Fig 11. Distribution of vsiRNA reads on the AV2 genome.
The normalised number of 21 nt reads (shown as one solid line per replicate; left y axis) and the proportion of all 21 nt reads mapping to this RNA (shaded and summed across replicates; right y axis) spanning each position in the viral genome across RNA1 (left), RNA2 (middle) and RNA3 (right). Both annotations are smoothed with a 100 nt sliding window. AV2 infected samples are shown in red (upper panels) and AV2-2st infected samples in blue (lower panels). Positive-sense reads are shown above the axis and negative-sense reads below the axis. Similar plots for other read lengths (22 nt, 24 nt, all reads) are shown in SR Fig. Note–as discussed previously–reads that map to the conserved 3′ regions may be assigned at random to one of the three segments, potentially leading to an apparent over-abundance in RNAs 1 and 2 due to mismapping of reads that actually derive from the more abundant RNA3.