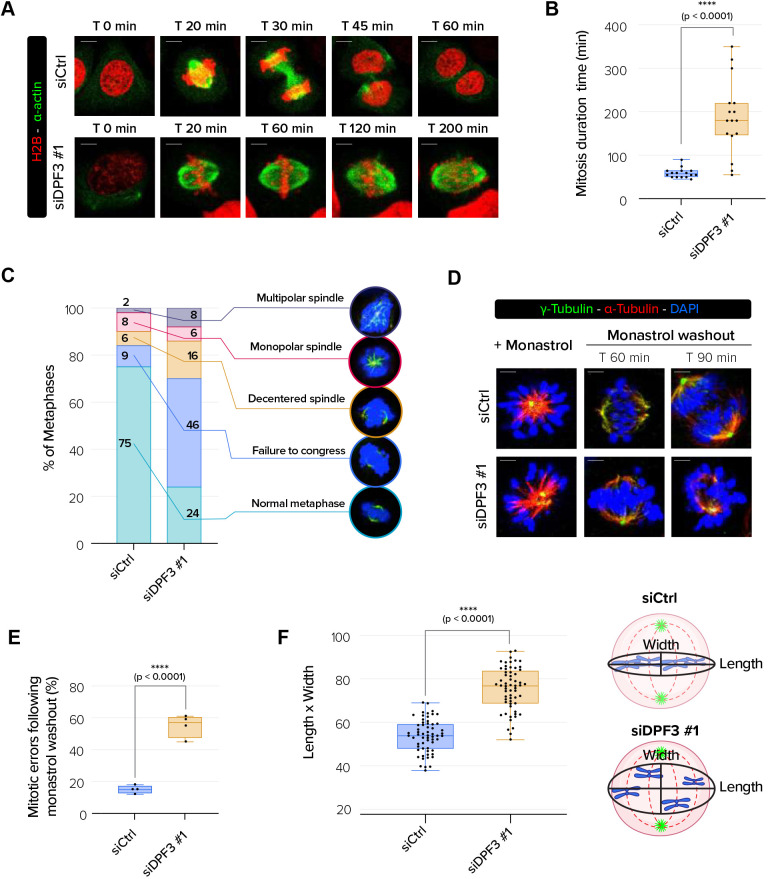
Fig. 5.
DPF3 is required for chromosome alignment. (A) Live-imaging experiments of H2B–RFP- and α-actin–GFP-expressing U2OS cells transfected with control siRNA (siCtrl) or DPF3 siRNA (siDPF3 #1) beginning 48 h post transfection. Scale bars: 5 µm. (B) Mitotic duration for H2AB–RFP- and α-actin–GFP-expressing U2OS cells transfected as in A (n=16 cells/condition from three independent experiments). (C) Quantification of the percentage of aberrant mitotic phenotype in U2OS cells transfected as in A (n=100 cells/conditions for one experiment). Representative images of abnormal mitotic phenotypes that were quantified are shown. (D) U2OS cells were transfected with control siRNA (siCtrl) or DPF3 siRNA (siDPF3 #1) and treated with monastrol (100 µM) for 3 h and then released in fresh culture medium. Cells were co-stained prior to monastrol washout and at 60 min or 90 min after washout. Mitotic spindles were labeled for α-tubulin (in red), γ-tubulin (in green) to mark centrosomes, and DAPI (in blue) to mark chromosomes. Merged channels with DAPI nuclear staining (in blue) are shown. Scale bars: 5 µm. (E) Quantification of the mitotic errors following monastrol washout from the experiment described in D (n=a total of 70 cells for each condition from three independent experiments). (F) Quantification of chromosome misalignment. The length/width ratio was determined as schematized in the right panel and calculated for each condition (n=20 cells/condition/experiment from three independent experiments). Boxes in B,E,F show the 25–75th percentiles, the line shows the median, and whiskers show the minimum and maximum values. P-values were calculated using unpaired two-tailed t-test (B,E,F). ****P<0.0001.