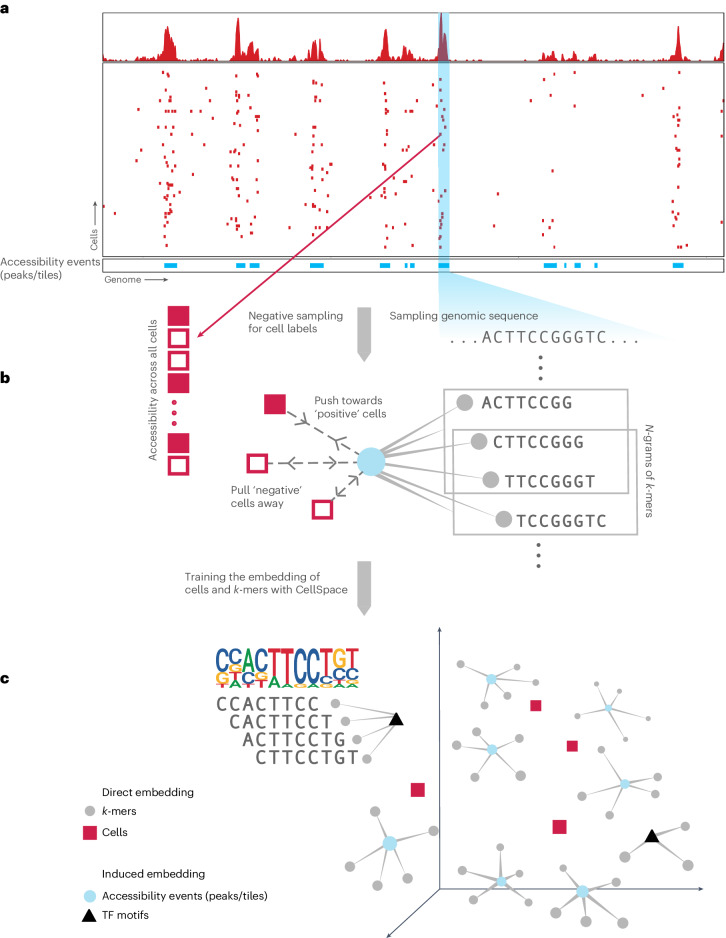
Fig. 1. CellSpace learns a sequence-informed embedding of cells from scATAC-seq.
Overview of the CellSpace algorithm. a, CellSpace samples sequences from accessible events (peaks or tiles) to generate training examples, each consisting of an ordered list of overlapping k-mers from the sampled sequence, a positive cell (where the event is open) and a sample of negative cells (where the event in closed). b, CellSpace learns an embedding of k-mers and cells into the same latent space. For each training example, the embeddings of the corresponding k-mers and cells are updated to pull the induced sequence embedding towards the positive cell and away from the negative cells in the latent space; learning contextual information, represented by N-grams of nearby k-mers, improves the embedding. c, Once the embedding of cells and k-mers is trained, TF motifs can be mapped to the latent space, allowing cells to be scored for TF activities based on TF-cell similarities.