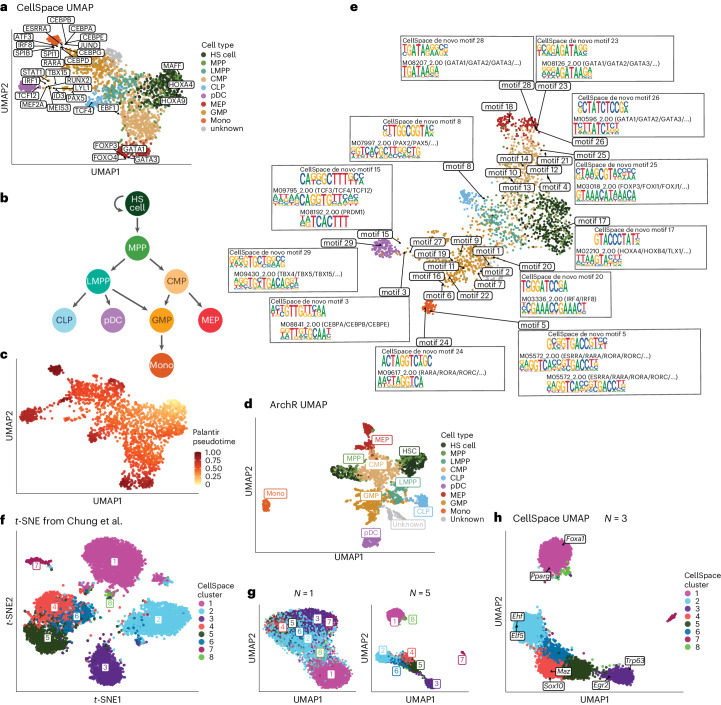
Fig. 2. CellSpace recovers latent structure and developmental hierarchies.
a, UMAP of CellSpace embedding for 2,154 cells from a small human hematopoietic scATAC-seq dataset annotated by fluorescence-activated cell sorting-sorted cell types. The embedding of key hematopoietic TF motifs is also shown. b, Current model of hematopoietic differentiation, with cell labels and colors as in a. c, Palantir pseudotime analysis using CellSpace embedding, with an HS cell starting point, identifies differentiation termini corresponding to CLP, pDC, GMP, MEP and monocyte (Mono) fates. d, UMAP of itLSI embedding based on cell-by-tile matrix using ArchR splits HS cell, MPP and MEP populations into two clusters due to batch effects. e, UMAP of cells and de novo motifs discovered based on the same trained CellSpace embedding as in a. DNA 10-mers that are frequent NNs of each cluster’s cells are identified and clustered by sequence content; 10-mer clusters are aligned and each converted to a PWM. f, Standard t-SNE from LSI dimensionality reduction of the cell-by-peak matrix for 7,846 cells from a murine fetal and adult mammary epithelial scATAC-seq dataset. The cells are annotated using CellSpace clusters (N = 3), and comparison with the original study was used to associate these clusters with cell types. g, UMAP of CellSpace embedding for the mouse mammary epithelial dataset shows the impact of N-gram parameter for N = 1 and 5. h, CellSpace with default N = 3 accurately captures developmental relationships between cell types. The key TF motifs in epithelial differentiation are also shown in the N = 3 CellSpace embedding.