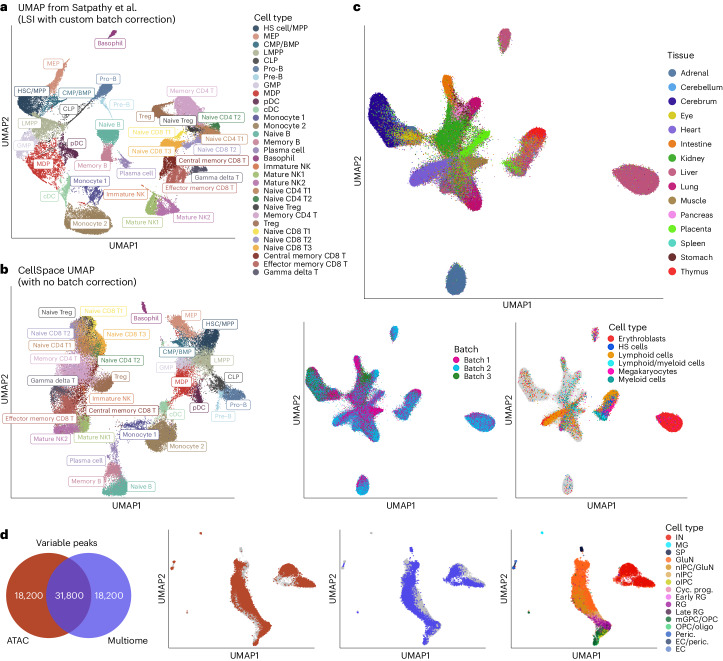
Fig. 4. CellSpace’s embedding implicitly mitigates donor- and assay-specific batch effects in large-scale scATAC-seq datasets.
a, UMAP of LSI dimensionality reduction with custom batch correction from original study of a large-scale multidonor human hematopoietic scATAC-seq dataset with 63,882 cells, annotated with major reported clusters. BMP, basophil–mast cell progenitor; MDP, monocyte–dendritic cell progenitor; cDC, conventional dendritic cell. b, CellSpace embedding of the large human hematopoietic dataset without any custom preprocessing recovers hematopoietic developmental hierarchy. c, UMAPs for CellSpace embedding of a human fetal tissue scATAC-seq atlas, with approximately 720,000 cells, labeled by tissue, by batch and by blood cell types across multiple tissues. d, CellSpace applied to human cortex chromatin accessibility data by joint embedding of two datasets: the scATAC-seq readout of the multiome dataset with 8,981 cells (Fig. 3a) and a (single-modal) scATAC-seq with 12,675 cells, processed with respect to their own peak atlases. The Venn diagram shows the top 50,000 most variable peaks from each assay, with 31,800 peaks in each atlas having nonzero overlap with the other atlas. The UMAP of the joint CellSpace embedding shows cells from each dataset, overlaid with cell type annotations from the original study. MG, microglia.