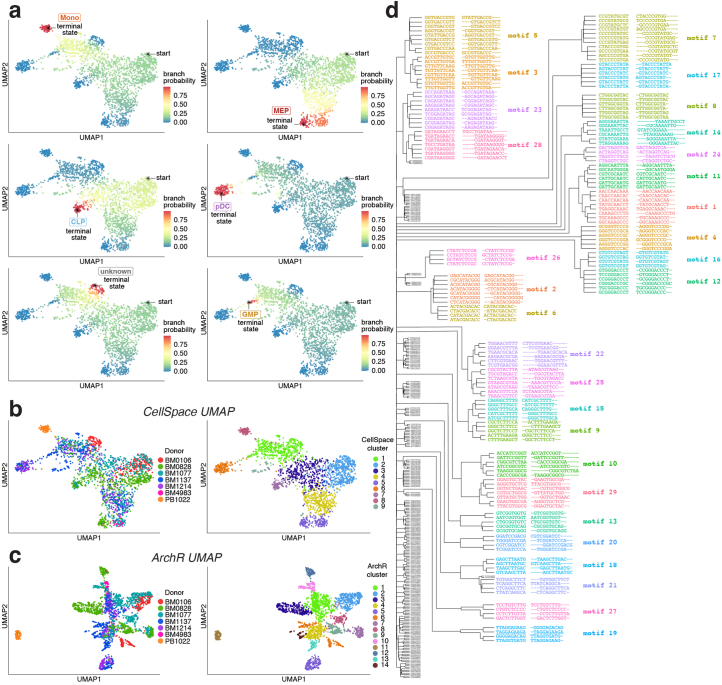
Extended Data Fig. 1. CellSpace recovers latent structure and developmental hierarchies.
a. Palantir branch probabilities showing trajectories to 6 termini in the small human hematopoietic dataset, including termini at CLP, pDC, GMP, an ‘unknown’ GMP-adjacent population, MEP, and monocytes. b. CellSpace embedding annotated by donor (left) and by Seurat’s SNN-based clustering (right), which largely recovers annotated cell types. c. ArchR embedding of the small human hematopoietic dataset annotated by donor (left) and by Seurat’s SNN-based clustering (right). d. Clustering of 10-mers retrieved as frequent nearest neighbors of cell clusters from the small human hematopoietic dataset; 10-mers in each cluster are aligned and then converted to PWMs of de novo CellSpace motifs (visualized in Fig. 2e).