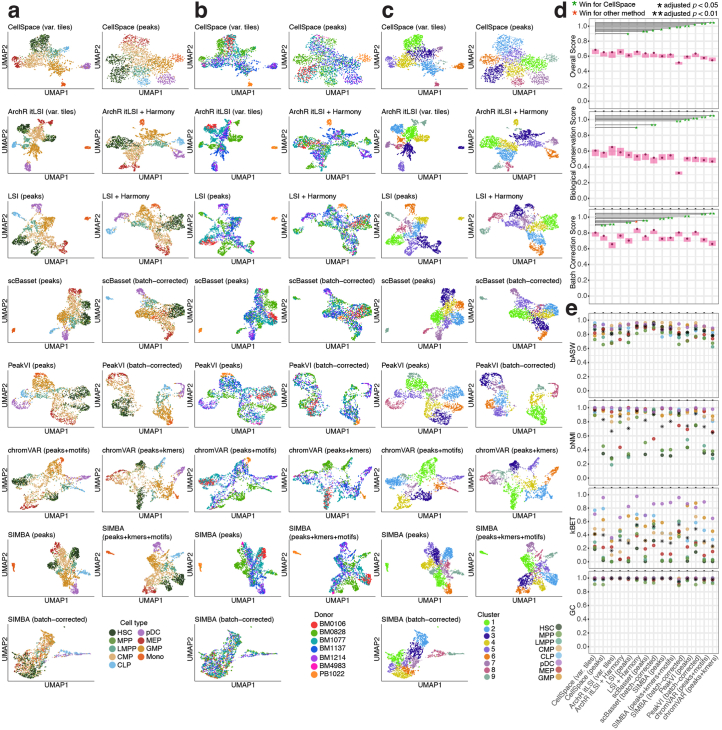
Extended Data Fig. 2. CellSpace outperforms other scATAC-seq embedding methods in batch mitigation/correction while preserving biological complexity.
a. UMAP visualizations for multiple scATAC-seq dimensionality reduction and embedding methods on the small human hematopoietic dataset, excluding the ‘unknown’ cell type. If the method offers an explicit batch correction option, the embedding corrected for donor batch effect is labeled as ‘batch-corrected’. b. UMAP visualizations annotated by donor (batch). c. UMAP visualizations annotated by Seurat’s SNN-based cluster. d. Performance metrics (aggregated biological conservation score, aggregated batch correction score, and overall score) for all methods on the small human hematopoietic dataset, excluding the ‘unknown’ cell type, with 95% confidence intervals over 1000 bootstrap samples. For each metric, all methods were compared in pairwise, two-sided tests on the bootstrapping samples, under the null hypothesis that the score difference is zero. The p-value for each comparison was computed using confidence interval inversion, and the values were FDR-adjusted across all comparisons. Only FDR-adjusted p-values comparing CellSpace to other methods are shown; *: adjusted p < 0.05; **: adjusted p < 0.01. e. Batch correction metrics reported per cell type, excluding the monocyte cell type which consists of a single batch. Average score over all cell types is also shown.