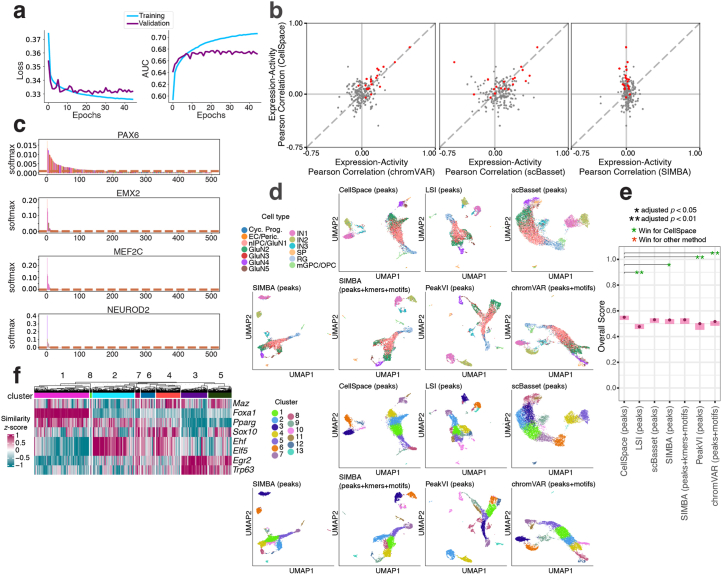
Extended Data Fig. 3. Single cell motif scoring using CellSpace accurately maps TF activities.
a. The scBasset model training converges after 40 epochs on the human cortex multiome dataset. b. Comparison of CellSpace vs. scBasset TF motif activity scores, CellSpace vs. SIMBA scores, and CellSpace vs. chromVAR scores based on correlation with gene expression in the human cortex multiome dataset. Important neurodevelopmental TFs shown in red. c. SIMBA motif scores for PAX6, EMX2, MEF2C, and NEUROD2 can be used to rank cells and learn an association with the top-ranked cell type. d. UMAP embedding and Seurat’s SNN-based clustering of the human cortex multiome dataset using multiple scATAC-seq embedding methods. e. Overall biological conservation score for all methods on the human cortex dataset (single batch), with 95% confidence intervals over 1000 bootstrap samples. For each metric, all methods were compared in pairwise, two-sided tests on the bootstrapping samples, under the null hypothesis that the score difference is zero. The p-value for each comparison was computed using confidence interval inversion, and the values were FDR-adjusted across all comparisons. Only FDR-adjusted p-values comparing CellSpace to other methods are shown; *: adjusted p < 0.05; **: adjusted p < 0.01. f. TF motif scores from the CellSpace embedding for the mammary epithelial dataset (embedding and clusters visualized in Fig. 2h).