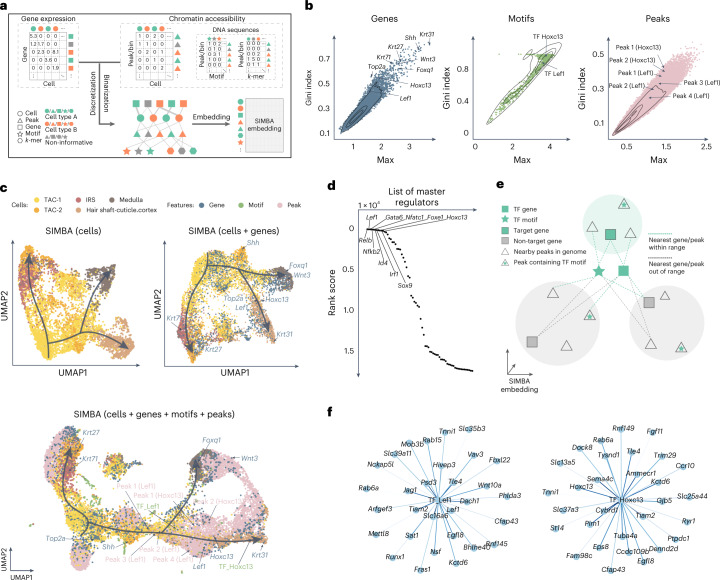
Fig. 4. Multimodal analysis of the SHARE-seq hair follicle dataset using SIMBA.
a, SIMBA graph construction and embedding in multimodal analysis. Overview of SIMBA’s approach to multimodal (scRNA-seq + scATAC-seq) data analysis. b, SIMBA metric plots of genes, TF motifs and peaks. All these features are plotted according to the Gini index against max score. Cell-type-specific genes, TF motifs and peaks are highlighted. c, UMAP visualization of SIMBA embeddings of cells (top left), cells and genes (top right) and cells along with genes, TF motifs and peaks (bottom). d, Ranked master regulators identified by SIMBA. The rank score indicates the rank of a given TF gene among all genes (including non-TFs) on the basis of the distance from the genes to the selected TF motif in the SIMBA embedding space. e, Schematic of SIMBA’s strategy for identifying target genes given a master regulator in the high-dimensional SIMBA embedding space (by default, 50 dimensions). The master regulator is represented by a TF motif (a star filled in green) and a TF gene (a square without borders filled in green). A colored shade indicates a SIMBA embedding area of a gene, containing a gene (a square with borders) and peaks (triangles) near the genomic locus of the gene. Green shades indicate areas of target genes, and gray shades indicate areas of non-target genes. f, Top 30 target genes of TFs Lef1 and Hoxc13, as inferred by SIMBA.