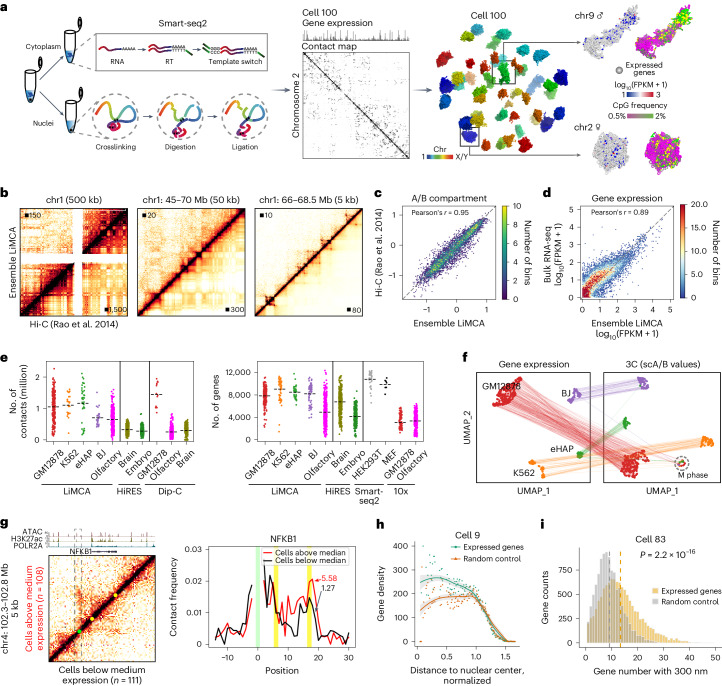
Fig. 1. Development of LiMCA.
a, Left, schematics of the LiMCA procedure. Right, the 20-kb resolution 3D genome structure of a representative cell; expressed genes are projected. RT, reverse transcription. b, Comparison of ensemble LiMCA and bulk Hi-C. The maximum intensity is indicated. c, Scatter-plot of the first eigen value between ensemble LiMCA and bulk Hi-C at 100-kb resolution. d, Scatter-plot of expression level (FPKM) between bulk RNA-seq (ENCODE ENCFF897XES) and combined expression profile of LiMCA. e, The median contact number of LiMCA (GM12878, 1.08 million, n = 220; K562, 1.14 million, n = 28; eHAP, 1.30 million, n = 42; BJ, 678,000, n = 32; olfactory, 652,000, n = 411) (left) is compared to HiRES (brain, 304,000, n = 399; embryo, 264,500, n = 300 (random sampled)) and Dip-C (GM12878, 1.45 million, n = 14; brain, 333,000, n = 300 (random sampled); olfactory, 252,000, n = 409). The median detected gene number of LiMCA (GM12878, 7,954, n = 221; K562, 9,806, n = 63; eHAP, 8,588, n = 42; BJ, 8,911, n = 63; olfactory, 4,528, n = 411) (right) is compared to HiRES (brain, 6,966, n = 399; embryo, 4,058, n = 300 (random sampled)), Smart-seq2 (HEK293T, 11,169, n = 35; MEF, 10,173, n = 7) and 10x chromium (olfactory, 3,556, n = 300 (random sampled of this study); GM12878, 2,754, n = 100 (random sampled)). f, Uniform Manifold Approximation and Projection (UMAP) embedding of four profiled cell lines based on gene expression profiles (left) or single-cell A/B values (right). The same cells are connected with lines. g, Contact matrices (left) around NFKB1, representing ensemble Hi-C data of NFKB1-high group (top left) and NFKB1-low (bottom right). Normalized contact frequency plot (right), centered at the NFKB1 upstream enhancer. The green and yellow dot/line indicates the position of the candidate enhancer and the transcription start site (TSS) and the transcription termination site (TTS) of NFKB1, respectively. h, Radial distribution of gene density; nucleus is sliced to 0.01 thickness. The error bands represent the 95% confidence interval (CI). i, Histograms show the distribution of cluster size within 300 nm for expressed genes and random control. Data were analyzed by a two-sided Mann–Whitney U-test.