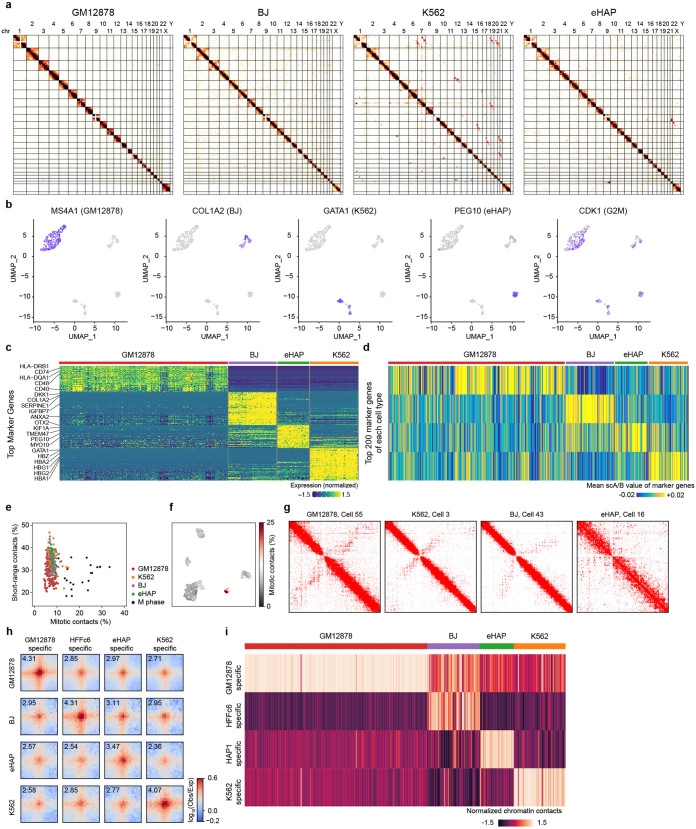
Extended Data Fig. 2. LiMCA accurately detects cell-type-specific gene expression and chromatin structures.
a, Ensemble contacts maps of four cell lines at 1 Mb resolution, translocations are highlighted with red arrow. b, UMAP showing four represented markers for each cell type and one maker gene of G2M phase. c, Expression of top cell-type-specific marker genes for each cell type, top 20 of each cell type are shown. d, Mean scA/B value of cell-type-specific marker genes among single cells. For each cell types, the top 200 marker genes were identified from the paired transcriptome data. e, Scatter-plot showing the mitotic contact band (2–12 Mb) ratio versus short-range contacts (< 2 Mb) ratio. f, UMAP visualizing the mitotic band ratio of cell line embedding. g, Represented contact maps of cells in metaphase cluster. h, Pile-up of cell-type-specific chromatin loops using ensemble interaction profiles from each cell type. i, Heatmap showing the enrichment of cell-type-specific chromatin loops among single cells.