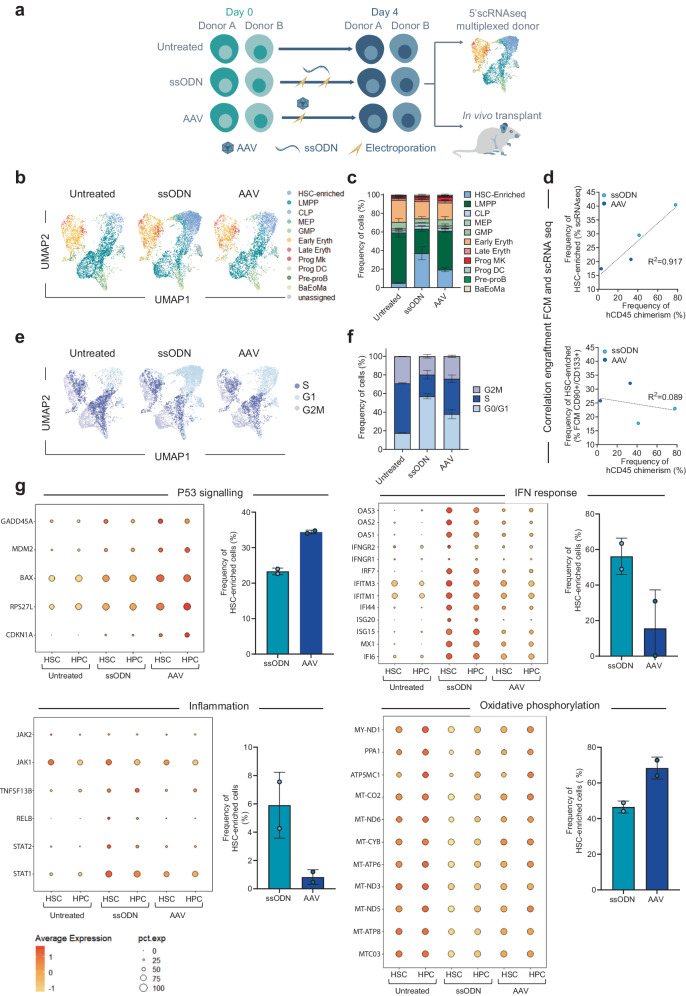
Fig. 3. Single-cell RNAseq characterization of the different editing protocols in plerixafor-mobilized HD HSPCs before in vivo injection.
a Representative schema and experimental design used for 5’scRNAseq analysis. b UMAP (Uniform Manifold Approximation and Projection) plot showing aggregated 5’scRNAseq data from non-edited and edited HSPCs analyzed the day of in vivo injection, n = 2 independent biological donors. Clusters and associated cell types (defined in the Methods section) are indicated by name and colors. c Average frequency ± SD of assigned cell types obtained out of n = 2 independent biological replicates coming from 2 donors. Two-way ANOVA followed by Bonferroni multi-comparison test. P-values are indicated. d Linear correlation between the frequency of HSC-enriched cells defined by scRNAseq data (upper plot) or by flow cytometry (FCM, lower plot) and the average frequency of hCD45+ chimerism obtained in BM 16–18 weeks after in vivo injection. Each dot represents data obtained from one HSPCs donor. For hCD45+ chimerism, the data from mice injected with the same donor were pooled, and the average frequency is shown. e UMAP plot showing aggregated 5’scRNAseq data illustrating the clusters and associated cell cycle phases of non-edited and edited HSPCs analyzed the day of in vivo injection. f Average frequency ± SD of cells associated with each cell cycle phase. e and f were plotted out of n = 2 independent biological replicates from 2 donors. Two-way ANOVA followed by Bonferroni multi-comparison test. P-value is indicated. g DotPlots from scRNAseq data representing gene expression profiles for selected relevant genes belonging to p53, IFN response, Inflammation, and oxidative phosphorylation pathways (rows) among hematopoietic stem cells (HSC) and the rest of progenitor cells (HPC). The size and the color of each dot represent the percentage of cells expressing a given gene and the average expression of the gene, respectively. On the side of each DotPlot is illustrated the average frequency ± SD of HSC-enriched cells (as defined in Fig. 3c) expressing selected relevant genes belonging to the p53, IFN response, Inflammation, and Oxidative phosphorylation pathways, for n = 2 independent biological replicates from 2 HSPC donors. Source data are provided as a Source data file.