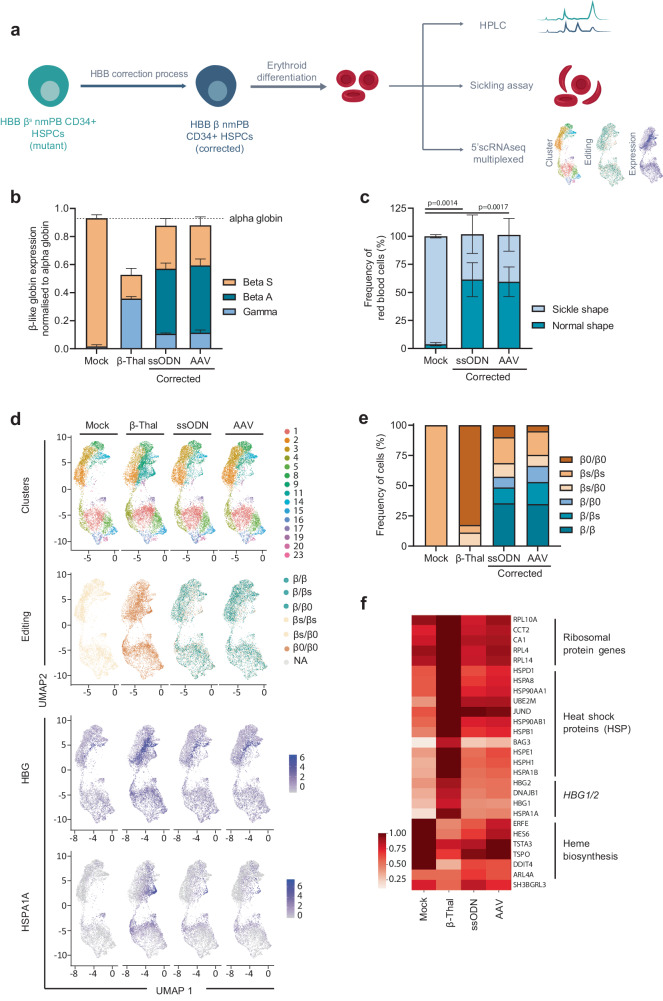
Fig. 5. Correction of the sickle phenotype in erythroid cells differentiated from edited HSPCs.
a Representative schema of gene correction protocol in HSPCs followed by erythroid differentiation and experimental design. b (RP) HPLC quantification of globin chains in erythroid cells derived from non-mobilized HSPCs from HbSS patients corrected with ssODN or AAV and compared to Mock and β-Thal controls. β-Like globin chains are normalized to α-globin chains. Data are expressed as average ± SD, for n = 3 independent biological experiments and donors. c In vitro sickling assay measuring the proportion of sickled RBCs under hypoxic conditions (0% O2). Plots represent the percentage of sickle- and normal-shape RBCs derived from Mock control, ssODN-edited, and AAV-edited cells. Data are expressed as average ± SD, for n = 3 independent biological experiments and donors. One-way ANOVA with Tukey’s comparison test. P-values are indicated. d UMAP (Uniform Manifold Approximation and Projection) plot comprising aggregated 5’scRNAseq data from differentiated erythroid cells analyzed 11- and 13-days post differentiation onset. Data represent the ssODN-edited and AAV-edited samples compared to Mock and β-Thal controls. Clusters determined by Louvain unsupervised clustering are indicated by different colors (Clusters), clusters representing HBB allelic genotype as determined by expressed HBB mRNA are indicated by different colors (Editing). Normalized expression of HBG and HSPA1A mRNAs are shown. The color gradient indicates the average expression of the indicated gene in each cell according to the scale shown. e Frequency of erythroid differentiated cells harboring each bi-allelic genotype as assigned in scRNAseq data for Mock, β-Thal, ssODN-edited, and AAV-edited samples. β = corrected allele, βs = sickle allele, β0 = Indel allele. f Heatmap showing expression (normalized expression values log transformed) of the top deregulated genes comparing Mock, β-Thal, ssODN-, and AAV-edited samples. General categories to which a list of genes belong are shown. Source data are provided as a Source data file.