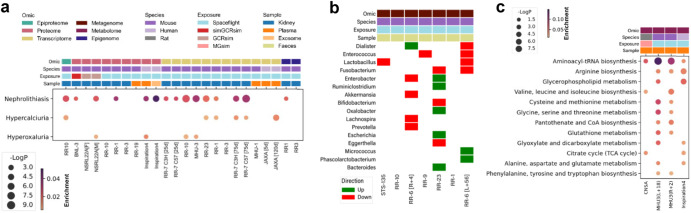
Fig. 4. Multi-mission pan-omic investigation of the contributors to nephrolithiasis risk during spaceflight.
a Multi-omic over-representation analysis of DisGeNET gene-disease associations related to nephrolithiasis. To integrate datasets from different omics modalities, species, missions and tissues, all biomolecules (e.g. phosphopeptides, proteins, transcripts and methylated DNA) were converted to the human orthologs where necessary and linked back to their HGNC gene symbol, aggregated and collapsed to single genes (e.g. multiple phosphosites, isoforms, CpG sites). An unadjusted, two-tailed -Log10(P-value) of 2 was considered significant for ontological term enrichment. b Categorical heatmap of differential abundance directionality in nephrolithiasis-related faecal microbial taxa after spaceflight. An unadjusted, two-tailed -Log10(P-value) of 1.3 was considered significant. c Over-representation analysis of KEGG module metabolic pathways with replication in at least two datasets. An unadjusted, two-tailed -Log10(P-value) of 1.3 was considered significant for ontological term enrichment. a, c Enrichment ratio; the number of differentially regulated hits in a dataset that belong to a given ontological term, normalised to the total number of statistically significant hits in the respective dataset.