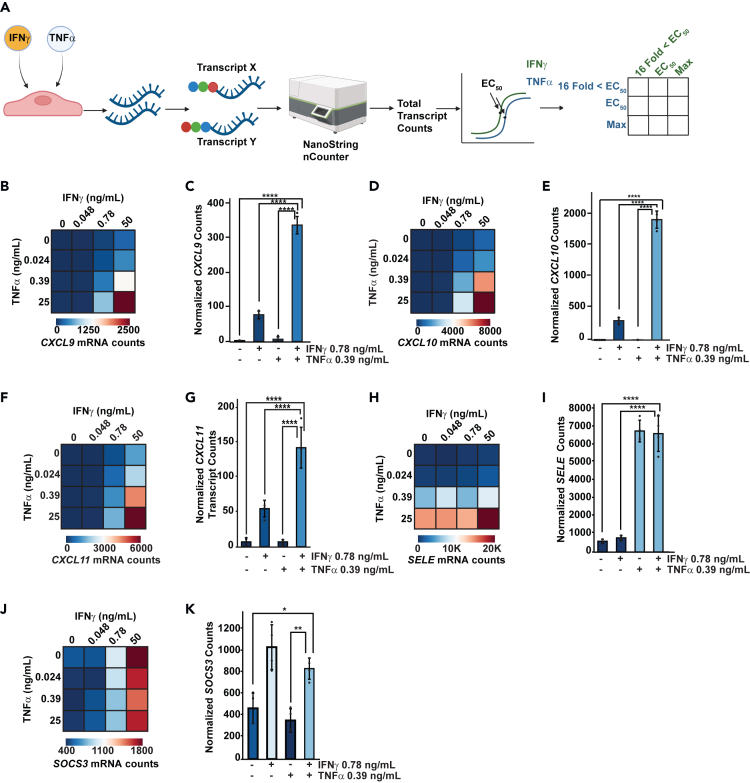
Figure 3.
Synergistic induction of proinflammatory genes is not dependent on saturation of IFNγ or TNFα signaling
(A) Schematic of NanoString workflow, identification of EC50 for IFNγ and TNFα (Figure S3A), and experimental design of dual-cytokine dose-response experiments.
(B, D, F, H, and J) Heatmap of CXCL9 (B), CXCL10 (D), CXCL11 (F), SELE (H), and SOCS3 (J) expression in response to maximal concentrations, EC50 and 16-Fold < EC50 of cytokine responsive genes of IFNγ and/or TNFα (n = 4 per condition).
(C, E, G, I, and K) Bar plot of normalized CXCL9 (C), CXCL10 (E), CXCL11 (G), SELE (I), and SOCS3 (K) transcript counts in Figures 2B–2D, 2F, 2H, and 2J. Bars indicate mean ± SD (n = 4 per condition). One-way ANOVAs were performed followed by Tukey’s post hoc analysis to determine statistical differences between treatment groups. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p ≤ 0.0001.