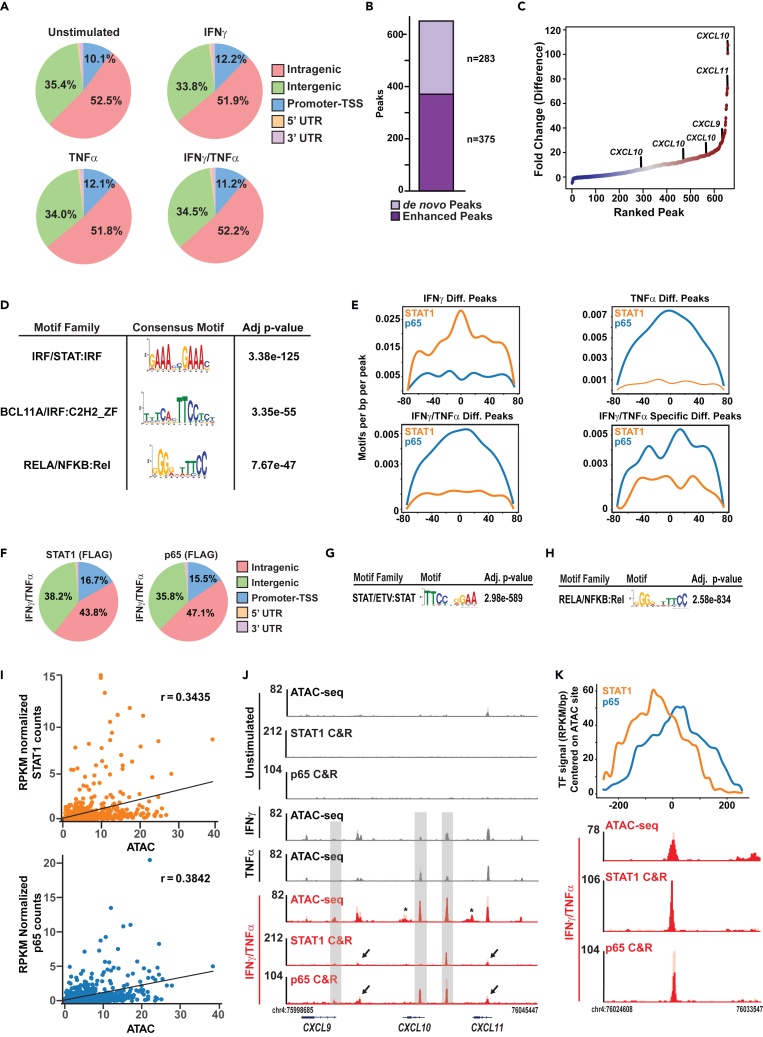
Figure 5.
Dual-cytokine stimulation results in the formation of de novo accessible elements at the CXCL9, -10, and -11 locus and recruitment of STAT1 and p65
(A) Pie chart of genomic localization of ATAC-seq peaks as a percentage of the total peaks. Unstimulated (upper left), IFNγ (upper right), TNFα (lower left), and IFNγ/TNFα (lower right) (n = 2 per condition).
(B) Stacked bar plot depicting number of enhanced (dark purple) or de novo (light purple) accessible regions in peaks differential to IFNγ/TNFα.
(C) Rank plot of log2(RPKM) ATAC-seq signal in SSAEs.
(D) Position weight matrix plots of the top motifs found in the IFNγ/TNFα-specific differential peaks.
(E) Signal plots of p65 or STAT1 motif density distribution in differential peaks in each cytokine condition.
(F) Pie charts of genomic localization of CUT&RUN peaks for FLAG-STAT1 and FLAG-p65. (FLAG) indicate the antibody used for CUT&RUN.
(G and H) Position weight matrix plots of the top motif found in the STAT1 peaks (G) and p65 peaks (H) in IFNγ/TNFα stimulated HAECs.
(I) Scatterplots of STAT1 signal (top, orange) and p65 signal (bottom, blue) vs. ATAC signal at SSAEs. R value indicates Pearson correlation coefficient.
(J) Gene tracks of ATAC-seq and CUT&RUN (C&R) for STAT1 and p65 at the CXCL9, -10, and -11 locus in HAECs at baseline and in response to IFNγ, TNFα or both (red). N = 2 per condition. Replicates are shown in two shades of color on the same track. Y axis is rpm/bp. Gray boxes indicate SSAEs co-bound by STAT1 and/or p65; asterisks indicate SSAEs with no TF co-binding; arrows indicate STAT1 and/or p65 binding at ATAC-seq sites that are not categorized as SSAEs.
(K) Gene track zoomed in on the CXCL10 and -11 intergenic enhancer showing the close proximity of STAT1 and p65 co-binding at this location centered on the ATAC-seq SSAE.