Figure 6.
Synergistically induced genes harbor transcriptional dependencies on p300/CBP and BET bromodomain-containing proteins
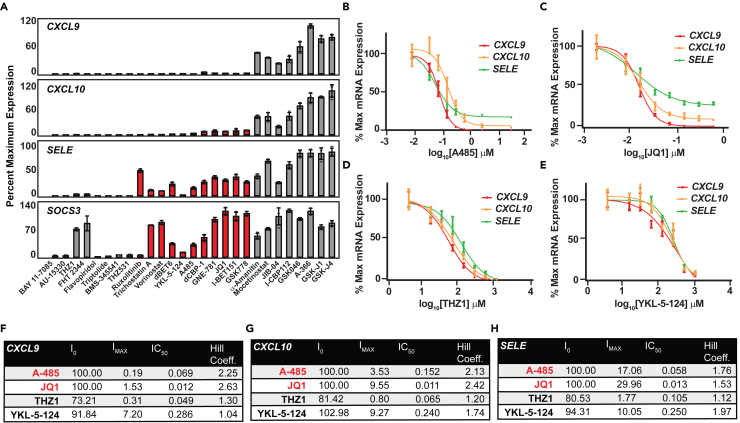
(A) Bar plots of gene expression of CXCL9 (top), CXCL10 (2nd from top), SELE (2nd from bottom), and SOCS3 (bottom) comparing untreated cells versus cells costimulated with IFNγ/TNFα+Vehicle or maximal concentrations (10μM) of transcriptional inhibitors. For AU-15330 and FHT1 samples were pretreated at 1 μM for 1 h before adding cytokine. Data shown are mean ± SD (n = 3 per condition).
(B–E) Dose-response curves for A-485 (B), JQ1 (C), THZ1 (D), and YKL-5-124 (E) in HAECs stimulated with IFNγ/TNFα and inhibitors (1 h). Graphs show % maximal expression (y axis) for CXCL9 (red line), CXCL10 (yellow line) and SELE (green line) vs. log10 inhibitor dose (x axis). Data are reported as mean ± SD (n = 4 per observation).
(F–H) Tables of biochemical parameters calculated from dose-response curves for each gene from (B–E).