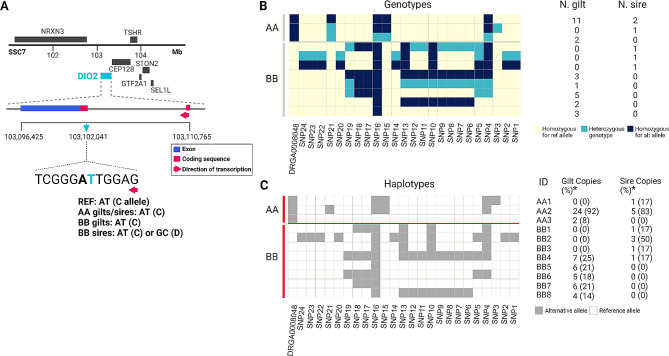
Fig. 1.
Identification of a missense variant (Asn91Ser) in porcine DIO2. (A) Sequenced exons are indicated in the context of DIO2 gene structure with neighboring genes on Sus scrofa chromosome (SSC7) 101–105 Mb region (Ensembl Release 105 (Dec 2021) used). Coding sequence (red colored box) encompasses the two sequenced exons (blue box). A single nucleotide variant site (light blue arrow) in the coding region on SSC7:103,102,041 is a missense variant changing asparagine (Asn) to serine (Ser) at position 91 of porcine DIO2 protein. This variant is present as a dinucleotide polymorphism with a synonymous variant (bolded left to the missense variant) on SSC7:103,102,040. AA or BB is genotype of DRGA0008048 (DRGA) SNP which is an intergenic variant ~ 14 Kb distal to DIO2. The alternative allele (GC) is labelled as D while the reference (REF) allele (AT) is C to distinguish the variant from DRGA. (B) Map of AA and BB genotypes for each SNP within the exonic region of DIO2 in gilts and sires used in trial-2. SNP labels correspond to labels in additional file 1; SNP24-SNP3, SNPs in the 3 prime untranslated region; SNP1 and SNP2, Asn91Ser variant and the neighboring synonymous variant. Each cell denotes genotype at each SNP site. Light yellow cells, homozygous genotype for reference (ref) allele. Sky blue cells, heterozygous genotype. Dark blue cells, homozygous genotype for alternative (alt) allele. Number of gilts and sires was shown for each row indicating a combination of all genotypes across SNP loci. (C) Haplotypes in the sequenced DIO2 region in AA versus BB genotype by DRGA SNP. The most probable haplotypes were determined by manual assignment. * denotes frequency of each copy of haplotypes within DRGA genotype (AA versus BB) and within parents (gilts versus sires). Gray cells, alternative allele. White cells, reference allele. Edited with BioRender