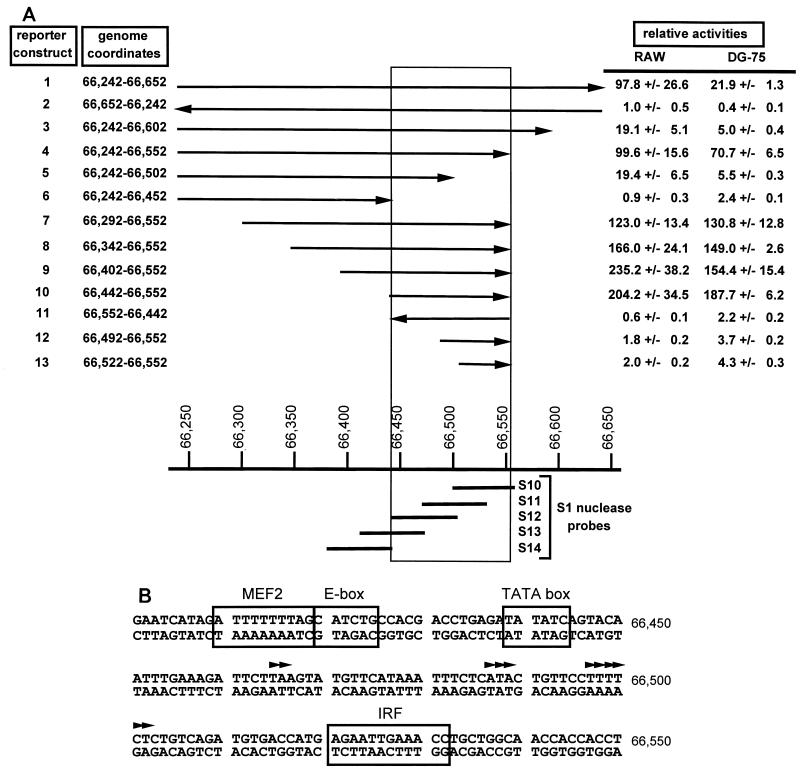
FIG. 4.
(A) Identification of the gene 50 promoter. The structures of γHV68 genomic fragments used to map the gene 50 promoter are shown, along with the genomic map coordinates. All viral genomic fragments were cloned upstream of the luciferase reporter gene in the pGL2 Basic vector (Promega, Madison, Wis.). The arrows pointing right indicate the genomic fragments that were cloned in the sense orientation, with the luciferase gene downstream of the putative gene 50 promoter, while the arrows pointing left indicate the genomic fragments that were cloned in the opposite orientation. The indicated reporter constructs (2 μg) were transfected into both the murine macrophage cell line RAW (RAW 264.7) and the human EBV-negative Burkitt's lymphoma B cell line DG-75. RAW cells were transfected with the lipid-based transfection reagent SuperFect according to the manufacturer's protocol (Qiagen, Santa Clara, Calif.). DG-75 cells were transfected with DEAE-dextran, as previously described (28). Cells lysates were prepared 48 h posttransfection and assayed for luciferase activity (7). The data were compiled from three independent experiments for each cell line, and the standard error of the mean is shown. Also shown are the positions of the single-stranded probes used in the S1 nuclease protection analyses (see text for discussion and Fig. 5). The boxed region denotes those sequences which, based on this analysis, are required for gene 50 promoter activity. Relative luciferase activity is the fold increase (mean ± standard error of the mean) over that observed with the parent pGL2 Basic reporter construct (Promega), which was assigned a relative activity of 1.0. (B) Computer analysis of the minimal gene 50 promoter for the presence of transcription factor binding sites. The gene 50 promoter sequenced was analyzed by MatInspector Professional software (23). IRF, interferon response factor; MEF2, myocyte enhancer binding factor 2; E box, binding site for the E family of bHLH transcription factors.