Figure 2. Interplay between H3.1 and DNA polymerases during replication.
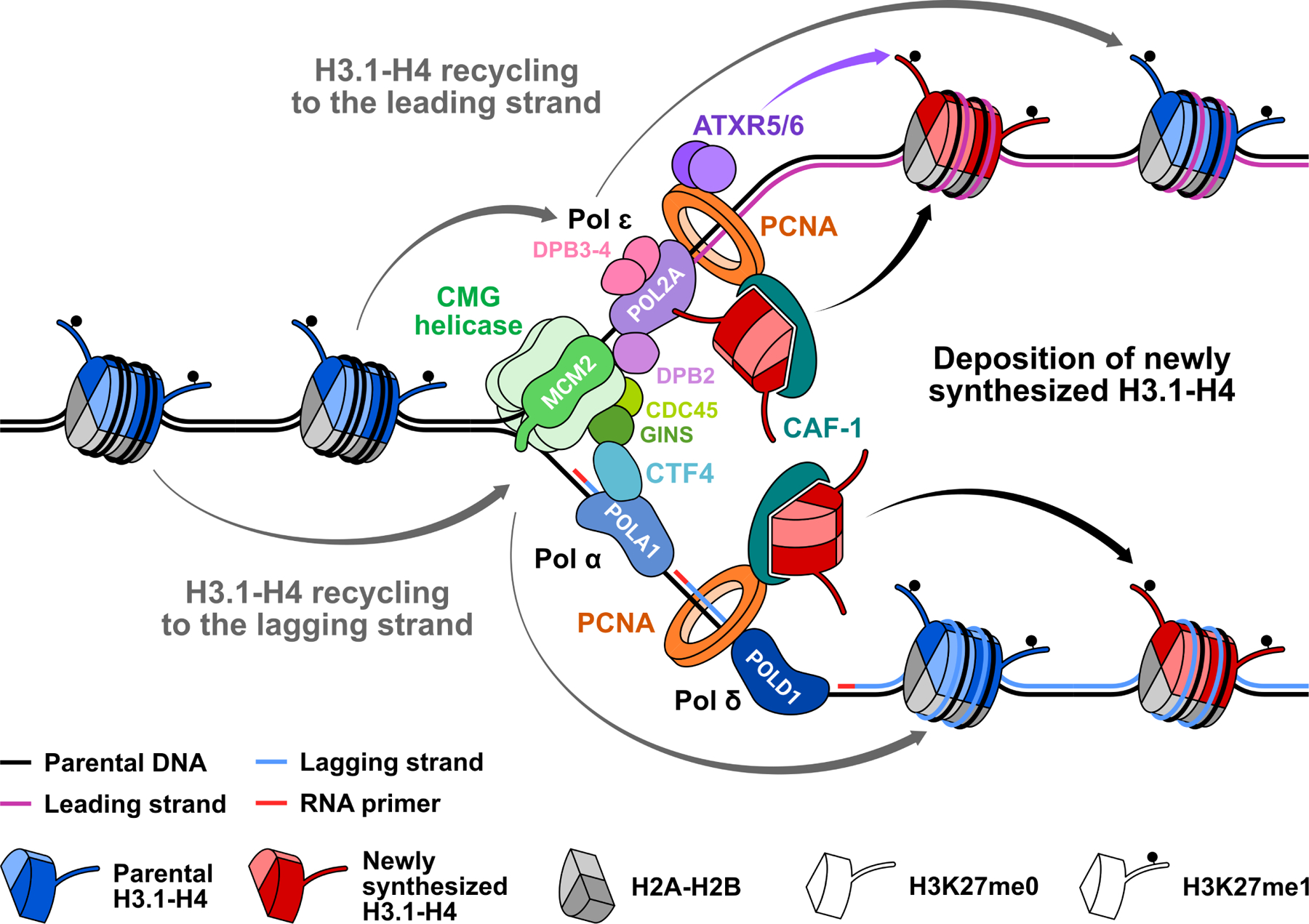
In constitutive heterochromatin, parental nucleosomes are enriched in H3.1 mono-methylated at K27. At the replication fork, the CMG helicase, which includes MCM2–7, CDC45, and GINS, unwinds parental double stranded DNA. DNA polymerase epsilon (Pol ɛ) then synthesizes the leading strand, while DNA polymerases alpha (Pol α) and delta (Pol δ) synthesize the lagging strand, with their respective catalytic subunits POL2A, POLA1, and POLD1. The CAF-1 complex interacts with the PCNA clamp to load newly synthesized H3.1-H4 tetramers on both strands of the fork, after which ATXR5/6 deposit H3K27me1. Parental H3-H4 are recycled symmetrically to the leading and lagging strands. POL2A can bind H3.1 in its unmethylated (i.e., newly synthesized) and mono-methylated (i.e., parental) forms with potential implications for epigenetic inheritance. DPB3–4 are the plant homologs of mammalian POLE3–4, two non-catalytic subunits of Pol ɛ that were shown mediate recycling of parental H3-H4 to the leading strand. The MCM2-CTF4-Pol α axis is required for recycling of parental H3-H4 to the lagging strand in mammals, and may accomplish similar functions in plants.
