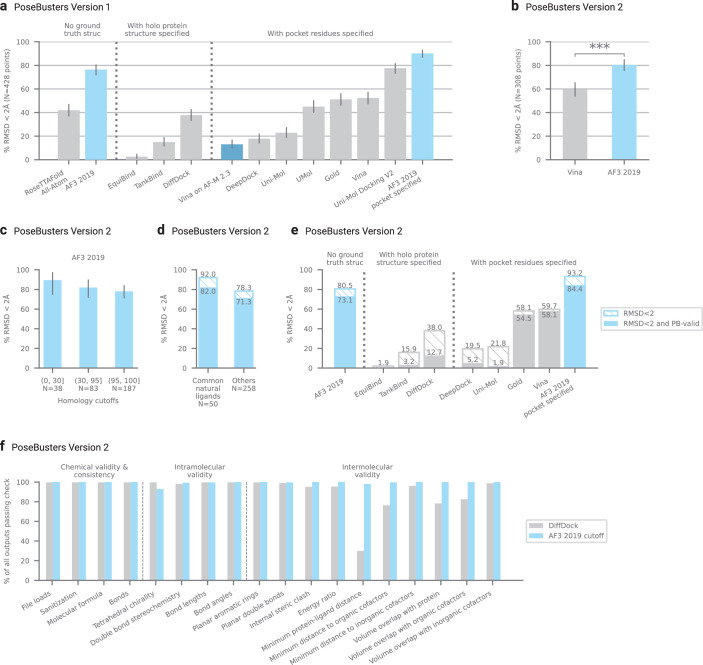
Extended Data Fig. 4. PoseBusters analysis.
a, Comparison of AlphaFold 3 and baseline method protein-ligand binding success on the PoseBusters Version 1 benchmark set (V1, August 2023 release). Methods classified by the extent of ground truth information used to make predictions. Note all methods that use pocket residue information except for UMol and AF3 also use ground truth holo protein structures. b, PoseBusters Version 2 (V2, November 2023 release) comparison between the leading docking method Vina and AF3 2019 (two-sided Fisher exact test, N = 308 targets, p = 2.3 * 10−8). c, PoseBusters V2 results of AF3 2019 on targets with low, moderate, and high protein sequence homology (integer ranges indicate maximum sequence identity with proteins in the training set). d, PoseBusters V2 results of AF3 2019 with ligands split by those characterized as “common natural” ligands and others. “Common natural” ligands are defined as those which occur greater than 100 times in the PDB and which are not non-natural (by visual inspection). A full list may be found in Supplementary Table 15. Dark bar indicates RMSD < 2 Å and passing PoseBusters validity checks (PB-valid). e, PoseBusters V2 structural accuracy and validity. Dark bar indicates RMSD < 2 Å and passing PoseBusters validity checks (PB-valid). Light hashed bar indicates RMSD < 2 Å but not PB valid. f, PoseBusters V2 detailed validity check comparison. Error bars indicate exact binomial distribution 95% confidence intervals. N = 427 targets for RoseTTAFold All-Atom and 428 targets for all others in Version 1; 308 targets in Version 2.