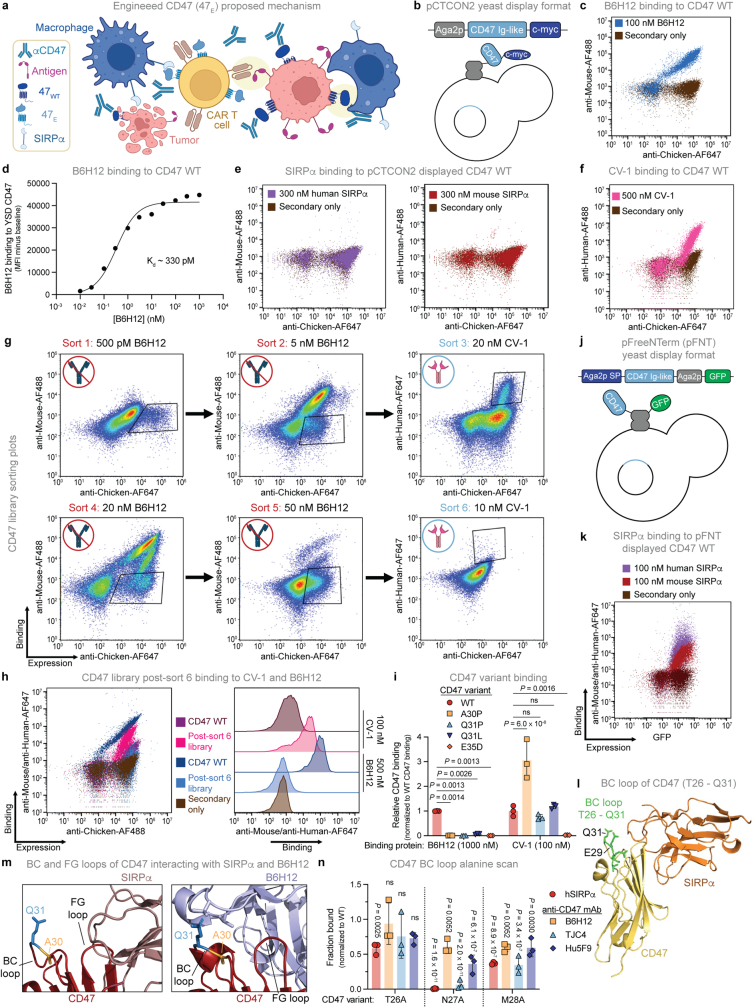
Extended Data Fig. 6. Engineered variants of CD47 retain SIRPα binding and demonstrate a loss of binding to some, but not all, anti-CD47 antibodies.
(a) Engineered CD47 (47E) mechanism: αCD47 antibodies bind tumour cells, but not 47E-T cells, triggering tumour-specific phagocytosis. The diagram was created using BioRender. (b) Cartoon of yeast displayed CD47 Ig-like domain using the pCTCON2 vector. CD47 is displayed as an N-terminal fusion. (c) Binding of 100 nM B6H12 to yeast displayed CD47 in pCTCON2 by flow cytometry. Representative of n = 3 independent experiments. (d) Binding curve of B6H12 to yeast displayed CD47 in pCTCON2, measured over multiple concentrations by flow cytometry. MFI of n = 1 experiment. (e) Binding of 300 nM human (top) and mouse (bottom) SIRPα to yeast displayed CD47 in pCTCON2 by flow cytometry. Data are representative of n = 3 independent experiments. (f) Binding of 500 nM CV-1 to yeast displayed CD47 in pCTCON2 by flow cytometry. Data are representative of n = 3 independent experiments. (g) Flow cytometry sorting plots of all six sorts of the CD47 library, indicating negative sorts to B6H12 and positive sorts to CV-1. Collected population indicated by the black box in each plot. (h) Binding of 500 nM B6H12 or 100 nM CV-1 to the yeast displayed CD47 library population collected after sort 6 or yeast displayed WT CD47. Data are representative of n = 2 independent experiments. (i) Binding of 1 μM B6H12 or 100 nM CV-1 to CD47 variants displayed on yeast using the pCTCON2 vector. Mean ± SD of n = 3 individual yeast clones, normalized to MFI from binding to 47WT. (j) Cartoon of yeast-displayed CD47 Ig-like domain using the pFreeNTerm (pFNT) vector. CD47 is displayed as a C-terminal fusion, along with GFP to monitor protein expression. (k) Binding of 100 nM human and mouse SIRPα to yeast displayed CD47 in pFNT by flow cytometry. Representative of n = 3 independent experiments. (l) Crystal structure of CD47 (yellow) binding SIRPα (orange) [PDB: 2JJS], identifying the CD47 BC loop (green), containing CD47 residues T26 – Q31. (m) Crystal structures of CD47 (red) binding SIRPα (dark pink, left) [PDB: 2JJS] and B6H12 (light blue, right) [PDB: 5TZU], identifying residues A30 (gold) and Q31 (blue), and the BC and FG loops of CD47. Structures are enlargements of the boxed regions in the full structures shown in Fig. 4c. (n) Binding of 100 nM B6H12, TJC4, Hu5F9, and hSIRPα to yeast displayed 47T26A, 47N27A, and 47M28A variants. Mean ± SD of n = 3 individual yeast clones, normalized to MFI from binding to 47WT. [(i), (n)] Two-way analysis of variance (ANOVA) test with Tukey’s multiple comparison test. ns = not significant. Comparison is between indicated group and binding to CD47 WT expressing cells.