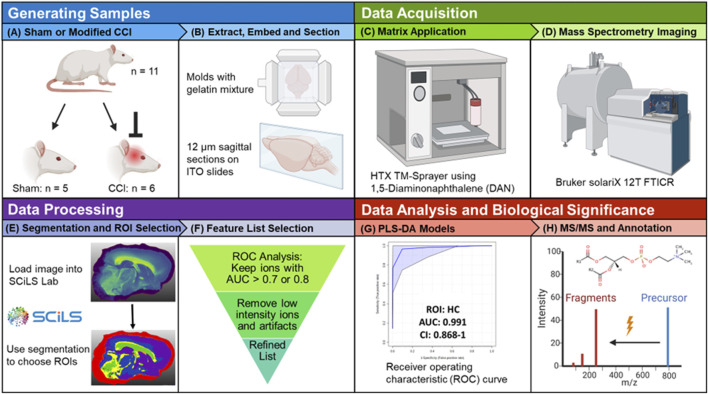
FIGURE 1.
Study workflow. (A) Male Sprague-Dawley rats were randomly assigned to a sham group (n = 5) or the three-impact rmTBI group (n = 6). (B) Rats were sacrificed 72 h post injury, the brains were extracted, separated into hemispheres, flash frozen, embedded in a gelatin mixture and sagittally-sectioned at 12 μm thickness. (C) ITO slides were sprayed with 8 passes of a 5 mg mL–1 DAN solution. (D) MALDI MSI data were collected on a Bruker solariX 12T FTICR mass spectrometer in positive ion mode. (E) Imaging data were loaded into Bruker SCiLS Lab software and the segmentation tool was used to select ROI. (F) ROC analysis was used to refine the lipid feature list for each selected ROI. (G) PLS-DA classification models for specific ROI were created in Metaboanalyst. (H) For annotation purposes, tandem MS experiments were performed on the discriminant species of importance in multivariate models. Created with BioRender.