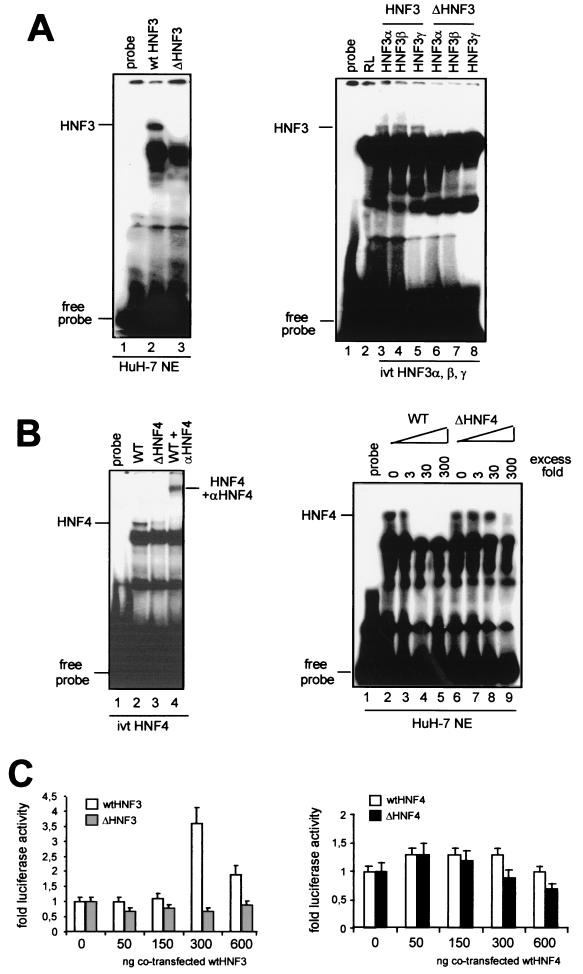
FIG. 3.
Mutations in the HBV enhancer I core domain decrease binding of HNF3 and HNF4. (A) 32P-labeled wt (left, lane 2) or mutant (left, lane 3) HNF3 oligonucleotides were used in gel shift experiments. The HNF3 oligonucleotides were incubated with HuH-7 nuclear extracts. The positions of the HNF3 complexes are marked. In vitro-translated (ivt) HNF3α, -β, and -γ proteins were incubated with wt (right, lanes 3 to 5) or mutant (right, lanes 6 to 8) HNF3 oligonucleotides. Incubation of reticulocyte (RL) extracts with wt HNF3 oligonucleotide is shown in lane 2. The positions of HNF3 complexes and free probe are depicted on the left. (B) HNF4 wt (left, lane 2) or mutant (left, lane 3) 32P-labeled oligonucleotides were incubated with in vitro-translated HNF4 protein. HNF4 was supershifted with polyclonal anti-HNF4 antibodies (left, lane 4). Competition experiments were performed with cold wt HNF4 (right, lanes 2 to 5) or mutant HNF4 (right, lanes 6 to 9) in the indicated molar excess in the incubation assay. (C) HuH-7 cells were transfected with wt, ΔHNF3, and ΔHNF4 enhancer I reporter gene constructs. Cotransfection experiments were performed with expression vectors for HNF3 (left) and HNF4 (right). The constructs were cotransfected with increasing amounts (0, 50, 150, 300, or 600 ng) of expression vectors for HNF3 (left) or HNF4 (right). The cells were harvested 48 h after transfection, and the luciferase activity was determined. The error bars indicate standard deviations. The activities of the samples in which only the luciferase reporter construct was transfected were set to 1. Note that dramatic changes were found in the basal activity with ΔHNF3 and -4 compared to that with the wt HBV construct (Fig. 2). The changes are shown as fold activation.