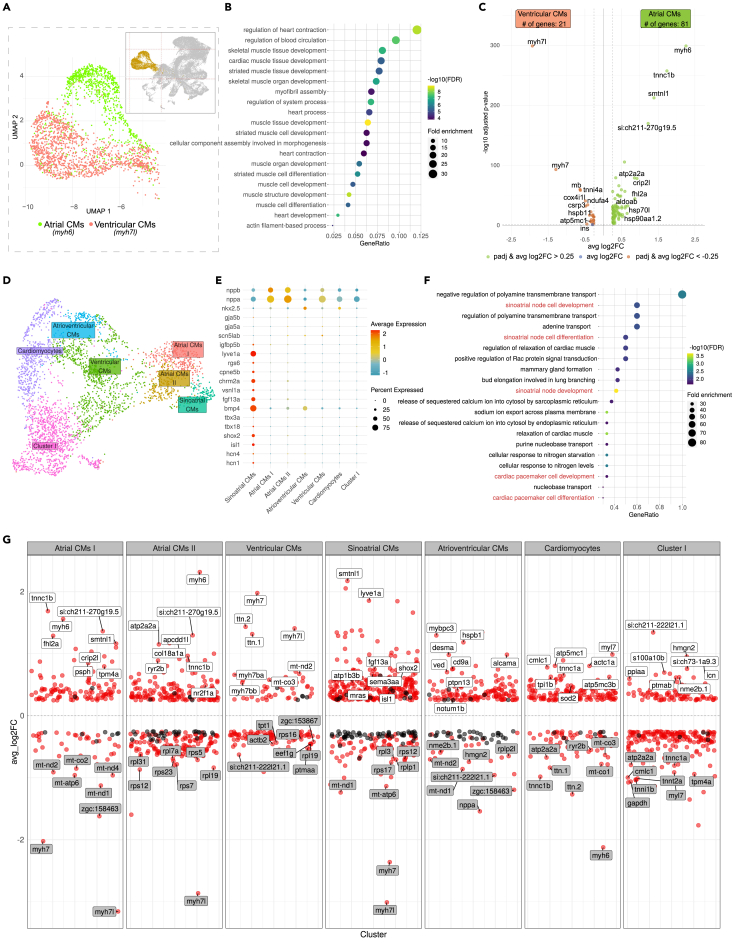
Figure 3.
Analysis of the “Myocardium” cluster revealed the diversity of myocardial cells and pinpointed sinoatrial cardiomyocytes within the atrial myocardium
(A) Two distinct identities of chamber myocardium could be delineated by molecular markers: Atrial CMs by expression of myh6 and Ventricular CMs by myh7l expression.
(B) Gene Ontology enrichment analysis of all genes enriched within the “Myocardium” cluster.
(C) Volcano plot depicting differentially expressed genes between atrial and ventricular CMs fractions.
(D) UMAP projection of re-clustered myocardial cells reflecting the heterogeneity of atrial and ventricular myocardium.
(E) Dotplot showing the expression (SCT normalized counts) of well-established gene signatures associated with working myocardium and sinoatrial pacemaker within “Myocardium” clusters.
(F) Gene Ontology enrichment analysis of genes enriched within the “Sinoatrial CMs” subcluster.
(G) Differentially expressed genes in each myocardial subclusters. Top 8 genes in terms of significance were labeled. Genes with adjusted p-value <0.05 are depicted as red dots while not significant genes (adjusted p-value >0.05) are shown in black. Source data: Tables S8, S9, and S10.