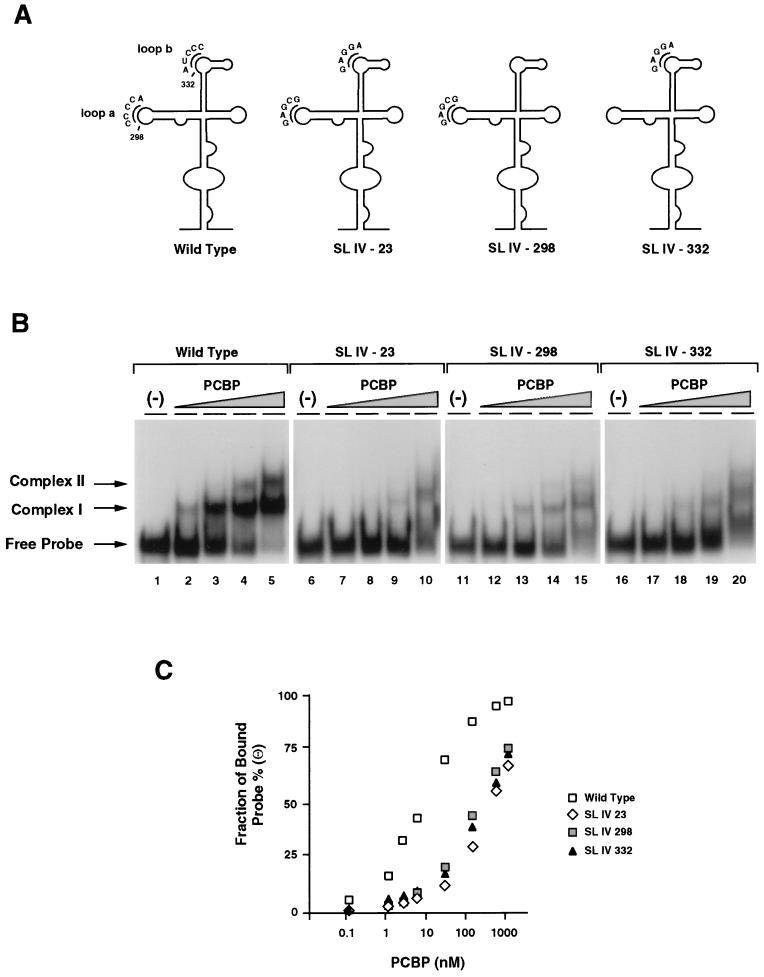
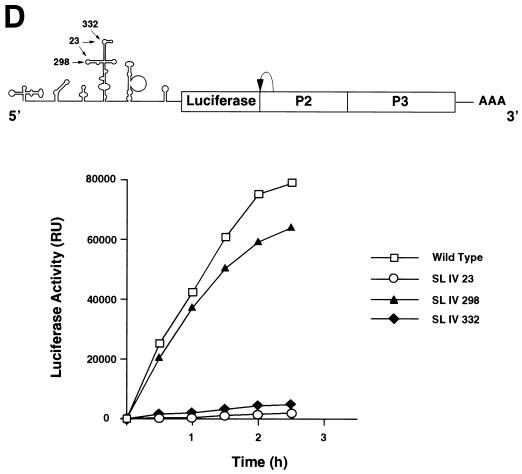
FIG. 2.
Effect of mutations within stem-loop IV on PCBP binding and viral translation. (A) Position and sequence of mutations within the predicted secondary structure of stem-loop IV. The C-rich loop a and loop b are indicated with their wild-type and mutated sequences: SL IV-23, SL IV-298, and SL IV-332. (B) RNA mobility shift analysis showing the effect of mutations within stem-loop IV probe on PCBP binding. RNA mobility shift experiments were performed with four different probes: wild-type stem-loop IV (lanes 1 to 5), SL IV-23 (lanes 6 to 10), SL IV-298 (lanes 11 to 15), and SL IV-332 (lanes 16 to 20). Each probe was incubated with buffer (−) or increasing concentrations of cellular PCBP (0.2 ng to 0.1 μg), as indicated at the top of the panel. Position of specific complexes I and II and the stem-loop IV probe (Free Probe) are indicated by arrows. (C) Binding affinity of PCBP to wild-type and mutated stem-loop IV RNAs. The fraction of radiolabeled probe bound (Θ) determined by mobility shift assays is plotted against PCBP concentration. (D) Translation efficiencies of wild type and stem-loop IV mutants in HeLa cells. At the top, a schematic representation of the poliovirus replicon carrying the luciferase reporter gene in place of the capsid proteins. The arrow indicates a recognition site for cleavage by the viral 2A protease. At the bottom, the translation of wild type and stem-loop IV mutants (SL IV-23, SL IV-298, and SL IV-332) is measured as luciferase activity and plotted as a function of the time after RNA transfections into HeLa cells.