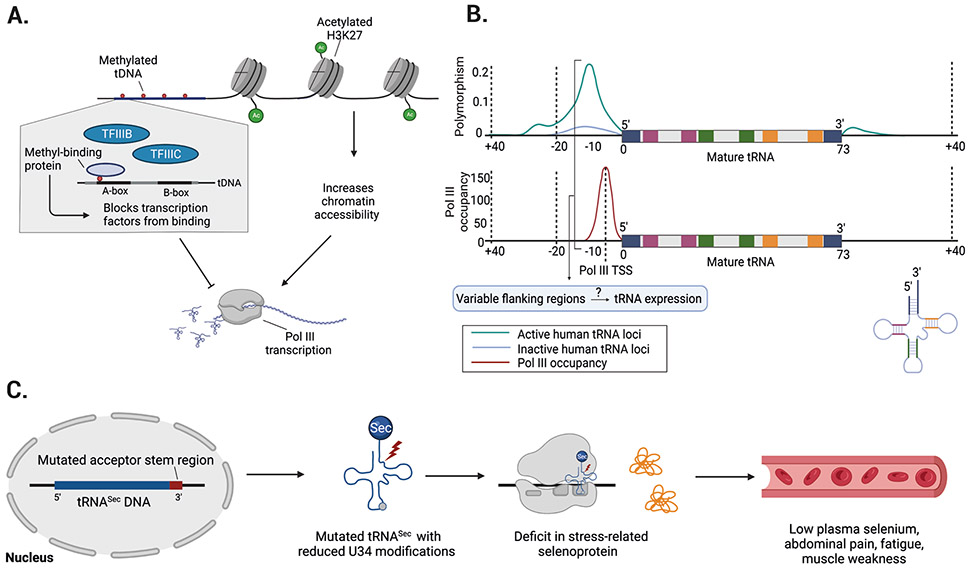
Figure 3. Epigenetic mechanisms and sequence variability might influence nuclear tRNA expression.
a ∣ Epigenetic mechanisms might regulate tRNA expression. For example, methylation of tRNA genes could block DNA polymerase III (Pol III) transcription. Similarly, histone marks (such as K3K27 acetylation) and overall nucleosome occupancy could influence the accessibility of chromatin at tRNA loci. b ∣ tRNA genes show high variability in their flanking regions and these hyper-variable regions69 overlap with Pol III occupancy and transcription start sites (TSSs)70. Top plot shows the frequency of single nucleotide polymorphisms (SNPs) for active and inactive tRNA loci. Bottom plot represents the distribution of Pol III occupation relative to each position within the tRNA gene and its flanking regions. The acceptor stem (blue), D-stem (red), anticodon stem (green), and T-stem (orange) are highlighted in the 2D tRNA structure for both plots. Flanking tRNA regions containing relatively high mutation rates can overlap with Pol III TSSs, which raises the important question of whether variable flanking regions influence tRNA expression. c ∣ Mutations in nuclear encoded tRNAs are rare, but one example is a mutation in the acceptor stem region (red) of the tRNASec gene that causes reduced U34 modifications and a deficit in stress-related translation of selenoproteins73. Patients with mutant tRNASec experience low plasma selenium levels, abdominal pain, fatigue, and muscle weakness. Part b is adapted from ref69 (top graph) and ref70 (bottom graph).