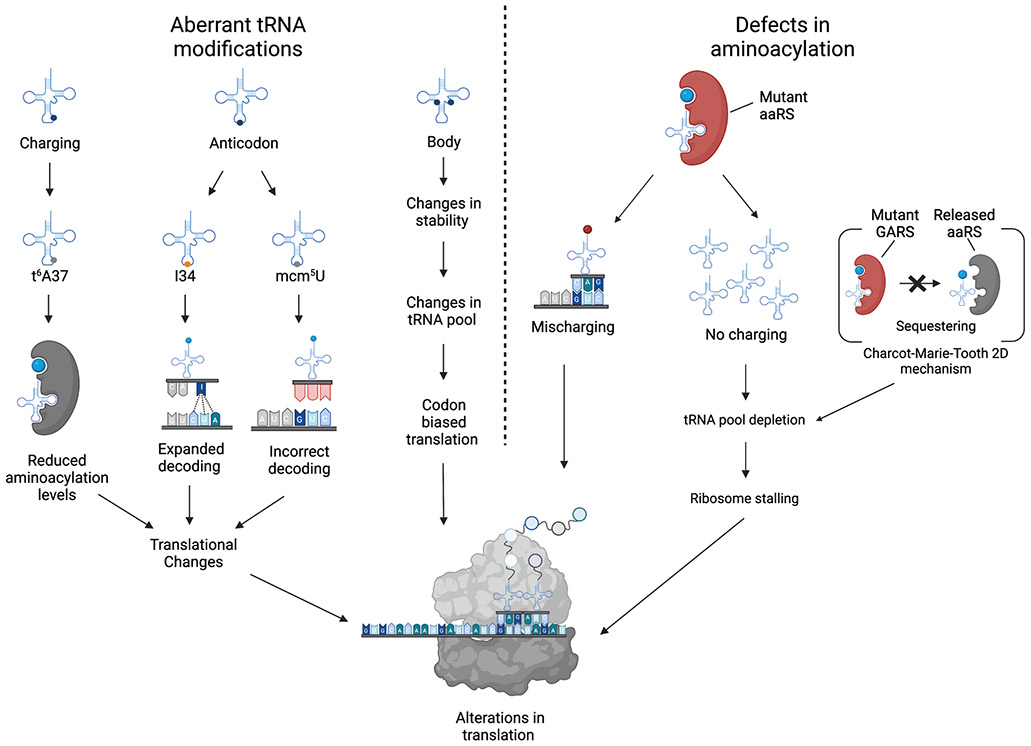
Figure 5. Defects in tRNA modifications or aminoacylation influence translation.
Translation elongation factors (principally eIF1α) capture aminoacylated tRNAs (with the exceptions of initiator tRNAMet, which requires eIF218, and tRNASec, which requires eIFSec19) and deliver them to ribosomes to be utilized in the synthesis of proteins. Inside the A-site of the ribosome, the tRNA recognizes cognate codon sequences by forming hydrogen bonds with its anticodon triplet at positions 34, 35, and 36. Codon–anticodon interactions in positions 35 and 36 follow Watson–Crick base pairing, whereas those at position 34 sometimes do not (wobble pairing)3. Thus, the canonical function of tRNAs as adapter molecules during protein synthesis is influenced by changes in tRNA modifications. Absence of the mcm5U modification in the anticodon results in incorrect decoding, whereas I34 modifications in the anticodon expand the tRNA decoding capabilities. Furthermore, tRNAs lacking t6A37 modifications leads to reduced aminoacylation levels. Moreover, defects in aminoacyl tRNA synthetases (aaRS) can cause mischarging (red circle), resulting in either incorporation of an incorrect amino acid or the absence of amino acid charging followed by tRNA degradation, depletion of the corresponding tRNA, and ribosome stalling at the cognate codon. The best-understood example is Charcot–Marie–Tooth disease type 2D, in which mutant glycyl-tRNA synthetase (GARS) fails to release bound tRNAGly, thereby sequestering it and leading to depletion of functional tRNAGly 105.