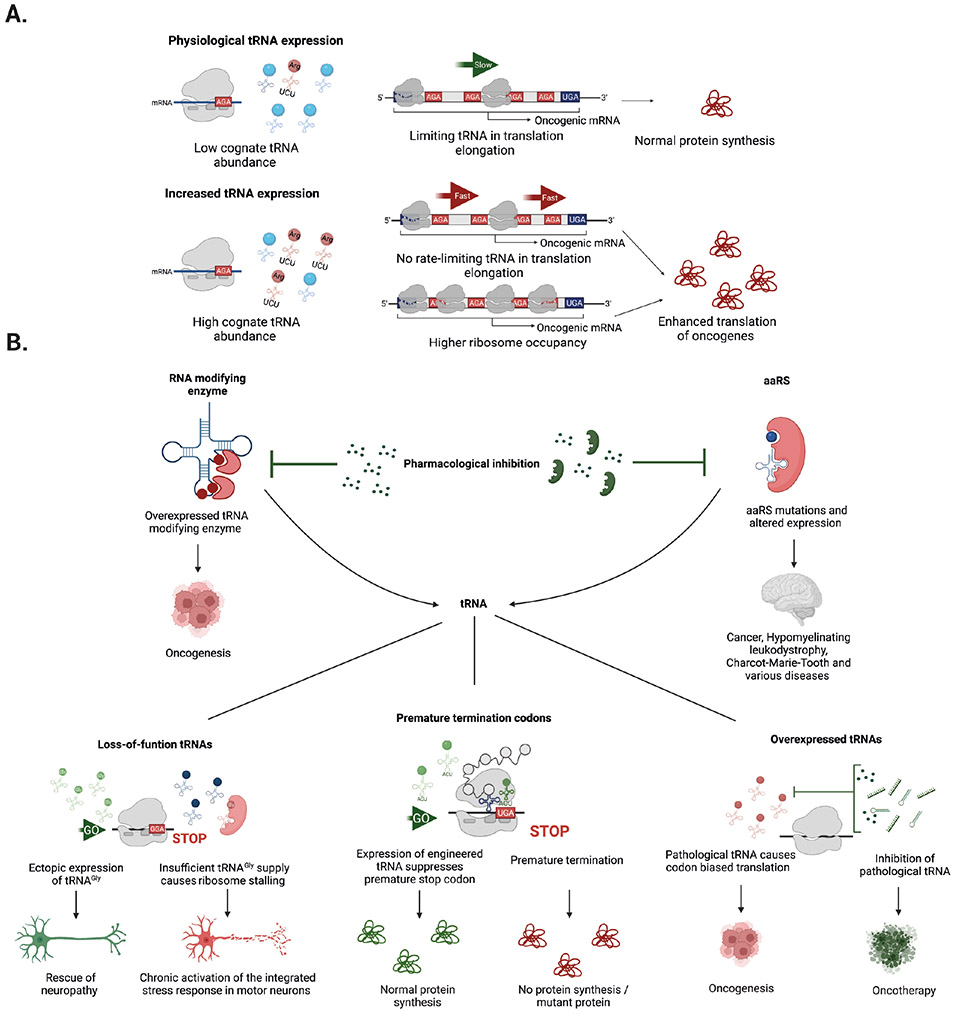
Figure 6. Alterations in the tRNA pool can drive disease and offer avenues for therapeutic intervention.
a ∣ Dysregulation of the levels of specific tRNAs leads to codon-biased translation. During steady state, the abundance of certain tRNAs could be a rate-limiting factor for the translation of oncogenic mRNAs enriched in their cognate codon (AGA highlighted in red), which restricts the translation rate and results in normal protein synthesis. Upregulation of the corresponding tRNA (red modified tRNAArg(TCT) versus other tRNAs (blue) removes this restriction, raising translation rates and increasing the production of oncogenic protein. This mechanism can result from increased ribosome occupancy or faster (enhanced) translation of mRNAs containing its cognate codon (red AGA). b ∣ Potential avenues for therapeutic intervention include pharmacological inhibition of tRNA modifying enzymes or aminoacyl tRNA synthetases (aaRSs) using small molecules or biological agents. These approaches could correct dysfunctional tRNA levels in cancer and various neurological diseases. Similarly, modulation of the expression of downstream tRNA effectors, either by ectopic restoration (green tRNA molecules) or inhibition of overexpressed tRNAs (red) might offer new strategies to treat diseases caused by altered levels of functional tRNAs. Potential approaches include pharmacological inhibition of pathological tRNAs by small molecules, RNA interference (RNAi), or antisense oligonucleotides (green). Another approach is the expression of anticodon engineered tRNAs to suppress premature termination codons and achieve normal protein synthesis.