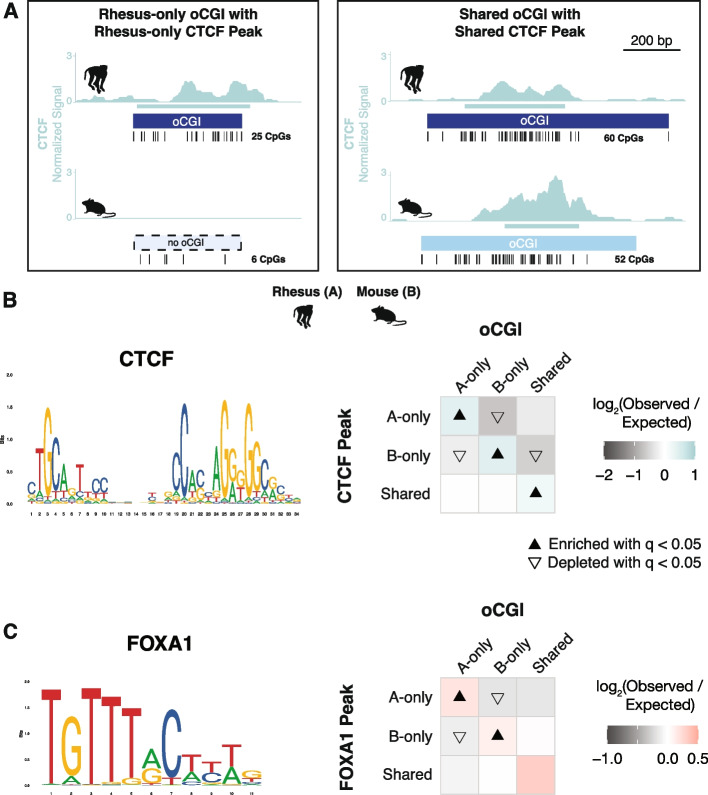
Fig. 6.
oCGI turnover is associated with changes in transcription factor binding. A Schematic illustrating how we compared species-specific oCGIs with species-specific transcription factor binding events in adult liver, using rhesus macaque and mouse as an example case. Left: a rhesus-only (species A-only) oCGI with a rhesus-only (species A-only) CTCF peak. Right: a shared oCGI with a shared CTCF peak. Ticks show the locations of CpG dinucleotides. B Left: the consensus motif for CTCF (MA1929.1 from the JASPAR database). Right: Enrichment and depletion in each indicated comparison of species-specific and shared oCGIs (top: A-only, B-only, Shared) and species-specific and shared CTCF peaks (left: A-only, B-only, Shared), compared to a null expectation of no association between oCGI turnover and peak turnover. Each 3 × 3 grid shows the results for a specific test examining oCGIs and their overlap with CTCF peaks. Each box in each grid is colored according to the level of enrichment over expectation (teal) or depletion (gray) of genome-wide sites that meet the criteria for that box. The color bar below each plot illustrates the level of enrichment or depletion over expectation. The filled upward-pointing triangles denote significant enrichment and open downward-pointing triangles denote significant depletion (q < 0.05, permutation test, BH-corrected; see Additional file 1: Fig. S22 and “Methods”). C Left: the consensus motif for FOXA1 (MA0148.1 from the JASPAR database). Right: Enrichment and depletion in each indicated comparison of species-specific and shared oCGIs (top: A-only, B-only, Shared) and species-specific and shared FOXA1 peaks (left: A-only, B-only, Shared). Shown as in (B) but with boxes colored according to the level of enrichment over expectation (red) and depletion (gray) of genome-wide sites that meet the criteria for that box