Abstract

The utilization of metallodrugs as a viable alternative to organic molecules has gained significant attention in modern medicine. We hereby report synthesis of new imine quinoline ligand (IQL)-based Cu(II) complexes and evaluation of their potential biological applications. Syntheses of the ligand and complexes were achieved by condensation of 7-chloro-2-hydroxyquinoline-3-carbaldehyde and 2,2′-thiodianiline, followed by complexation with Cu(II) metal ions. The synthesized ligand and complexes were characterized using UV–vis spectroscopy, TGA/DTA, FTIR spectroscopy, 1H and 13C NMR spectroscopy, and pXRD. The pXRD diffractogram analysis revealed that the synthesized ligand and its complexes were polycrystalline systems, with nanolevel average crystallite sizes of 13.28, 31.47, and 11.57 nm for IQL, CuL, and CuL2, respectively. The molar conductivity confirmed the nonelectrolyte nature of the Cu(II) complexes. The biological activity of the synthesized ligand and its Cu(II) complexes was evaluated with in vitro assays, to examine anticancer activity against the MCF-7 human breast cancer cell line and antibacterial activity against Gram-positive (Staphylococcus aureus) and Gram-negative (Escherichia coli and Pseudomonas aeruginosa) bacterial strains. The CuL complex had the highest cytotoxic potency against MCF-7 breast cancer cells, with an IC50 of 43.82 ± 2.351 μg/mL. At 100 μg/mL, CuL induced the largest reduction of cancer cell proliferation by 97%, whereas IQL reduced cell proliferation by 53% and CuL2 by 28%. The minimum inhibitory concentration for CuL was found to be 12.5 μg/mL against the three tested pathogens. Evaluation of antioxidant activity using 2,2-diphenyl-1-picrylhydrazyl revealed that CuL exhibited the highest antioxidant activity with IC50 of 153.3 ± 1.02 μg/mL. Molecular docking results showed strong binding affinities of CuL to active sites of S. aureus, E. coli, and estrogen receptor alpha, indicating its high biological activity compared to IQL and CuL2.
1. Introduction
Globally, the burden of cancer is expected to rise from 19.9 million new cases in 2022 to 21.3 new million cases in 2025,1 suggesting a growing burden of cancer in the society.2 By 2040, it is anticipated that the number of cancer cases worldwide will exceed 28 million annually.3 Among all the cancers, breast cancer is the most commonly diagnosed type and the leading cause of cancer death in women globally.4 Currently available cancer drugs, including platinum-based metallodrugs, such as cisplatin, are frequently associated with significant side effects and development of drug resistance, resulting in treatment failure.5,6 Drug resistance is also a challenge for treatment of infections, where emergence of resistant strains reduces efficacy of antimicrobial drugs, resulting in rising morbidity and mortality caused by infectious diseases.7,8 Consequently, there is an urgent need to develop new anticancer and antimicrobial drugs with novel mechanisms of action to overcome drug resistance,9 increase efficacy, reduce side effects, and improve overall treatment outcomes for patients. These outcomes may be achieved through the development of various metallodrugs, considering that metal-based coordination complexes have unique reactivity, electronic features, and redox chemistry.5,6
In coordination complexes, the metal–ligand interactions play a significant role in drug design and development as they serve as bridges between medicinal compounds, cells, and/or pathogenic organisms, thereby dictating their biological activities.10,11 These activities are mostly due to the variable oxidation states, coordination geometries, kinetic characteristics, and organic ligands linked to the metal center, which make metals and metal complexes interesting candidates for therapeutic research.11,12 Research trends for developing metal-based chemotherapy agents are currently toward nonplatinum-based metal complexes involving copper, titanium, gallium, germanium, palladium, gold, cobalt, ruthenium, tin, etc.13 because of their fewer side effects than platinum-based medications.14 Copper metal ions are important in cell division and growth and accumulating research evidence supports its potential utility in the battle against cancer.15 Copper-based complexes have also been studied for their direct free radical scavenging abilities, demonstrating their potential to mitigate the harmful effects of oxidative stress.16,17 Copper can also enhance the effectiveness of some chemotherapeutic drugs like bioactive organic ligands by sensitizing cancer cells. This synergistic effect can improve the overall efficacy of cancer therapy.18
Imine-based ligands, like quinolines and N-donor metal complexes, have generated significant interest due to their fascinating chemical properties and potential biological uses, including antibacterial, and anticancer effects.19,20 Quinolines are versatile heterocyclic aromatic compounds containing nitrogen and various moieties.21 They are present in both synthetic and naturally occurring biologically active substances, making them crucial in pharmaceutical research for development of novel drugs.22 Since quinolines have great potential to function as bioactive reagents on their own, their transition-metal complexes can further increase their biological activity.20−23 Quinoline motif Schiff bases, such as (E)-N-(furan-2-yl methylene) quinoline-8-amine and their metal complexes with Mn(II), Co(II), Ni(II), Cu(II), and Zn(II), have also been reported to possess antimicrobial, antioxidant, and in vitro cytotoxicity activities.24 The studies on the structure–function relationship also indicate that altering the ligand and the general structure of the complexes significantly increases the potency and efficiency of the metal complexes for various biological applications.25
In this work, we synthesized Cu(II) complexes from imine quinoline derivative ligand. The synthesized complexes underwent comprehensive evaluation for their in vitro cytotoxicity, antibacterial, and antioxidant properties following standard procedures. Additionally, in silico molecular docking studies were conducted to predict the interaction and binding energies of the compounds with key amino acids, including those from Staphylococcus aureus, Escherichia coli, and estrogen receptor alpha. Furthermore, density functional theory (DFT) and time-dependent (TD)-DFT calculations were employed to explore the UV–visible spectra and quantum mechanical descriptors of the complexes.
2. Materials and Methods
2.1. Chemical and Materials
Absolute ethanol (99.9%, Finkem), 2,2′-thiodianiline (98%, Japan), 7-chloro-2-hydroxyquinoline-3-carbaldehyde, acetic anhydride 99.8%, acetic acid glacial 99.5%, dimethylformamide (DMF, 99%), phosphorus oxychloride (POCl3, 98%,) methanol (99.5%), n-hexane (99%), dichloromethane (98%), ethyl acetate (99.5%), chloroform (99%), copper chloride dihydrate (CuCl2·2H2O, 98%), l-ascorbic acid (99%), dimethyl sulfoxide (DMSO) (99%), and 2,2-diphenyl-1-picrylhydrazyl (DPPH) were used for synthesis of the ligand and metal complexes, and for testing. All the chemicals and reagents were of analytical grade.
2.2. Characterization of the Synthesized Ligand and the Metal Complexes
2.2.1. Analysis of 1H NMR and 13C NMR Spectroscopy
The 1H and 13C NMR spectra of the ligand were acquired using an Avance 400 spectrometer (Bruker, Bremen, Germany) with TMS as internal standard. The spectrometer operates at 400 MHz, DMSO, for 1H-NMR and 100 MHz, DMSO, for 13C-NMR at 298 K.
2.2.2. Fourier Transform Infrared Spectroscopy Analysis
The infrared absorption spectra of the synthesized ligand and the Cu(II) metal complexes were recorded using an IR affinity-1 Fourier transform infrared (FTIR) spectrophotometer (Shimadzu). The spectrophotometer operated in the range of 4000–400 cm–1, with an accumulation of 32 scans and a resolution of 4 cm–1. Pure KBr discs served as the standard, while the samples were prepared as KBr pellets with a sample-to-KBr ratio of 1% w/w.
2.2.3. Analysis of the Electronic Absorption Spectra
The electronic absorption spectrum of the synthesized IQL and its Cu(II) complex solutions in DMSO were analyzed using a Shanghai Drawell Scientific Instrument spectrophotometer. The spectra were recorded in the wavelength range of 200–800 nm.
2.2.4. Powder X-Ray Diffraction Analysis
The crystallinity of the ligand and the Cu(II) complexes were studied using X-ray diffractograms (XRD patterns) produced with a Rigaku Ultima IV X-ray diffractometer utilizing Cu K radiation with a generator at 40 kV and 30 mA at 25 °C temperature (PD-M5-10 min-WAXS Prog-Div 15–80°). At a rate of 2 min per minute, the diffraction angle (2θ) range for XRD spectroscopy was reached, ranging from 0 to 90°.
2.2.5. Thermal Analysis
Using the Thermal Analysis Instrument of the SDT-Q600 analyzer (TA Instruments), the thermal stability of the Cu(II) complexes was examined. This device produced differential thermal analysis (DTA) and synchronized thermogravimetry analysis (TGA) curves. An alumina sample container was filled with a sample mass of roughly 7.0 mg, and a heating rate of 10 °C min–1 was used while an airflow of 100 mL min–1 was in place.
2.3. Synthesis of the IQL and Its Cu(II) Complex
2.3.1. Synthesis of the Imine Quinoline Derivative Ligand: (Z)-3-(((2-((2-Aminophenyl) Thio)phenyl)imino)methyl)-7-chloronaphthalen-2-ol
The ligand (Z)-3-(((2-((2-aminophenyl) thio) phenyl)imino)methyl)-7-chloronaphthalen-2-ol was synthesized,26 as described in Scheme 1, by reacting 7-chloro-2-hydroxyquinoline-3-carbaldehyde and 2,2′-thiodianiline in 1:1 molar ratio specifically, to a solution of 7-chloro-2-hydroxyquinoline-3-carbaldehyde (10 mmol, 1.5 g) in 15 mL ethanol, 2,2′-thiodianiline (10 mmol, 2.2 g) in 15 mL ethanol was gradually added while stirring. Then, the solution was stirred for 1–2 h at room temperature. As a result, a yellow solution was obtained. Then, 5–10 drops of glacial acetic acid were added to this mixture while stirring. The reaction completion was monitored using thin-layer chromatography. Finally, the yellow precipitate formed at the bottom was filtered and washed using 20 mL of ethanol. After drying, the actual yield of the product was determined.
Scheme 1. Synthesis of IQL, (Z)-3-(((2-((2-Aminophenyl) Thio)phenyl)imino)methyl)-7-chloronaphthalen-2-ol.
Yellow solid: % yield = 89.2, MP 145–150 °C, MW = 405.9 g/mol. The major IR peaks (cm–1): (vNH) 3854, 1734; (OH) 3742; v(C=N) 1601, 1560, and data of 1H NMR and 13C NMR are described in Table S1. Scheme 1 presents the synthetic route of the ligand.
2.3.2. Synthesis of the Cu(II) Complexes
Cu(II) chloride dihydrate (CuCl2·2H2O; 0.170 g; 1 mmol) was added to an ethanolic solution of (Z)-3-(((2-((2-aminophenyl) thio) phenyl)imino)methyl)-7-chloronaphthalen-2-ol (0.220 g; 1 mmol) for the synthesis of CuL in a 1:1 molar ratio, and (CuCl2·2H2O; 0.170 g; 1 mmol) and IQL (0.440. g; 2 mmol) for the synthesis of CuL2 in 1:2 molar ratio of metal ion to ligand. In both cases, the solution mixture was stirred and refluxed at 60–70 °C for 3–4 h. The reddish and green solutions obtained for CuL and Cul2 were cooled and the resulting precipitates were filtered out, washed with cold ethanol, and dried.26 Yield = 69.23% for CuL and 75.4% for CuL2, respectively.
2.4. Evaluation of Biological Activities
2.4.1. Antioxidant Activity Analysis
The antioxidant activities of the IQL, CuL, and CuL2 were assessed using a conventional protocol.27 Separate test tubes were prepared with standard ascorbic acid and various concentrations of IQL, CuL, and CuL2 (ranging from 50 to 250 μg/mL for each) from a 50 mg/mL stock solution of IQL, CuL, and CuL2. To each prepared solution, 3 mL of a 0.1 mM methanol solution of DPPH was added. The test tubes were then placed in the dark and allowed to incubate for 20 min. Following incubation, the absorbance of the solutions at 517 nm was measured using a UV–vis spectrophotometer against methanol as a blank. Ascorbic acid (ranging from 0.2 to 1.0 mg/mL) in methanol was used as a standard. The experiments were conducted in triplicate, and the antioxidant activity was determined as the scavenging percentage activity using eq 1.
| 1 |
where Ac is the absorbance of standard/control and As is the absorbance of IQL, CuL, and CuL2 solutions.
2.4.2. Antibacterial Activity Analysis
The antibacterial activities of IQL and its Cu(II) complexes were tested on Mueller Hinton agar media using an agar-well diffusion method27 following standard protocol. Specifically, 1 mL of actively developing clinical strains of Gram-negative bacteria [E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 248 (P. aeruginosa)] and Gram-positive [S. aureus ATCC 25926], containing about 1 × 107 CFU mL–1, 0.5 McFarland Standard, was dispersed on a homogeneous plate using cotton swabs separately. Wells were created in each agar culture medium, using a sterile cork borer (6 mm in diameter) and filled in with the samples having a concentration of 50 μg/mL. DMSO served as the negative control, while gentamicin was utilized as the positive control. After preparation, the culture media were refrigerated for 30 min before being placed in an incubator at 37 °C for 24 h. The diameter of the inhibition zones was then measured using a Vernier caliper in millimeters to ascertain the zones of inhibition.
2.4.2.1. Minimum Inhibitory Concentration
A microplate dilution method with slight modification28 was used to determine the minimum growth inhibition concentration (MIC) for the Cu(II) complexes and IQL. Solutions of copper complexes and the ligand were prepared in 10% DMSO in 2-fold dilutions. Bacterial suspensions were prepared from Mueller–Hinton broth and 100 μL of the suspensions were added to 1.5 μL microcentrifuge tubes with 300 μL of copper complexes and ligands. Microcentrifuge tubes containing sample solutions without bacterial suspension were used as negative controls, whereas those containing bacterial suspension with various gentamicin concentrations as positive controls. The tubes were incubated for 24 h at 35 ± 2 °C. Using a UV–vis spectrophotometer (Drawell, DU-8800R), absorbance at 630 nm was used to measure the antimicrobial activity. The negative control was employed as a blank.29
2.4.3. In Vitro Cytotoxicity Test on the Breast Cancer Cell Line
MCF-7 breast cancer cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 2 mM l-glutamine. The cells were maintained at 37 °C in a humidified 5% CO2. In vitro cytotoxicity was evaluated using the 3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay kit (Cell Proliferation Kit I MTT, Roche) according to the manufacturer’s instructions. The cells were cultured in a transparent, flat-bottomed 96-well microplate at a density of 5 × 103 cells/well. Cells were then treated in triplicate wells with 2-fold dilutions of compounds IQL, CuL, CuL2 and cis-platin (positive control) diluted with growth media at concentrations ranging from 6.125 to 200 μg/mL. Untreated control cells were incubated with matched solvent concentrations prepared in growth media. The cells were treated for 24 h and in the last 4 h of the experiment, an MTT labeling reagent was added to the cells at a concentration of 0.5 mg/mL. DMSO was used to solubilize formazan crystals, and optical density was measured at 570 nm wavelength.
2.4.4. Molecular Docking
Essentially, the binding modalities of ligand interaction at the active sites of the proteins were predicted from molecular docking calculations30 using the Autodock vina31 program package. The proteins were downloaded from the Protein Data Bank (http://www.rcsb.org/pdb) and used for the molecular docking investigations against S. aureus (PDB: 2w9h), E. coli (PDB: 6F86), and estrogen receptor alpha (ERα; PDB: 5GS4). Before docking, the proteins were repaired by a series of processes. Among these steps were the elimination of coligands and solvent molecules, the addition of hydrogen, chain repair, and active site selection. The compounds that were tested also went through an energy minimization using DFT calculations prior to the molecular docking.32 Different conformers were analyzed to ascertain the stability of H-bonds, π–alkyl, π–Sigma, π–π, π–anion, and van der Waals adducts. Using Discovery Studio software, the conformers with the lowest binding free energies were used to visualize how the compounds interacted with the active amino acids.33
2.4.5. DFT Calculations
DFT calculations employing B3LYP34−36 hybrid functional together with 6-311++G(d,p) basis set37 for the light atoms and the Los Alamos National Laboratory 2-double-ζ (LanL2DZ) pseudo-potential for the copper atom38 were employed using the Gaussian 16 program package.39 The polarizable continuum solvation model in its integral equation formalism together with DMSO as a solvent was used to account for solvent effects. Dispersion correction40 was employed to account for nonbonding interactions. Vibrational frequency calculations were performed to ensure that the optimized geometries are real minima without imaginary frequencies (Table S7). The TD-DFT was used to calculate absorption energies at the same level of calculations.
Quantum chemical descriptors were calculated at the same level of theory. The highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO) energies were used to predict the quantum mechanical descriptors: band gap (Eg = ELUMO – EHOMO), electronegativity [χ = −1/2 (EHOMO + ELUMO)], electronic chemical potential (μ = 1/2 (EHOMO + ELUMO) = −χ), global chemical hardness [η = 1/2 (ELUMO – EHOMO)], global softness (σ = 1/2η), global electrophilicity index (ω = μ2/2η), and nucleophilicity index (Nu = 1/ω) of the compounds.41,42
2.5. Statistical Analysis
The Statistical Analysis Software (SAS 14.1) was utilized to examine the mean antioxidant and antibacterial characteristics of the IQL and its Cu(II) complexes using one-way ANOVA with a 5% probability.
3. Results and Discussion
3.1. Physical Properties
The synthesized IQL, CuL, and CuL2 were characterized using physicochemical and spectroscopic methods. These compounds were observed to be colored, odorless, and relatively stable at room temperature. While the ligand was soluble in organic solvents such as dichloromethane, methanol, ethanol, acetonitrile, DMF, DMSO, and chloroform, the complexes exhibited solubility in these organic solvents but were insoluble in water. Additionally, the molar conductivity (ΛM) values (recorded at 10–3 M concentration) of the complexes in DMSO solutions were determined and are presented in Table 1.
Table 1. Molar Conductivities and Chloride Test of the Cu(II) Complexes.
| compounds | color | molar conductance (S cm2 mol–1) | chloride test |
|---|---|---|---|
| IQL | yellow | ||
| [CuL(H2O)2Cl2] | brick red | 4.45 | no white precipitate |
| [CuL2Cl2] | green | 11.05 | no white precipitate |
3.1.1. Molar Conductivities and Chloride Test
The compounds were dissolved in DMSO for molar conductance assessment, aiming to determine the electrolytic and nonelectrolytic attributes of the synthesized metal complexes. The molar conductance values of 4.45 and 10.05 S cm2 mol–1 were recorded for CuL and CuL2, respectively (Table 1). Accordingly, the complexes were of nonelectrolytic nature. In addition, a chloride test was carried out using the silver nitrate salt to check the presence of chloride ions in the complex structure. The absence of white precipitate confirmed that there are no chloride ions in the outer sphere of the Cu(II) coordination. Thus, the geometries of [CuL(H2O)2Cl2] and [CuL2Cl2] have been proposed to be octahedral, with the chemical formulas for CuL and CuL2 being C22H20Cl3CuN3O3S and C44H32Cl4CuN6O2S2, respectively.
3.2. 1H NMR and 13C NMR Spectroscopy Analysis Result of the Ligand 1H NMR Spectral Analysis
The proton NMR of the ligand was obtained in DMSO-d6, which is displayed in (Table S1 and Figure S1). The IQL exhibits a singlet peak in the range of δ 9.30 ppm as a result of the hydroxyl presence.43 The IQL exhibits a singlet at δ 8.90 ppm due to the imine (HC=N) group’s proton.44 These changes show imine chelation with metal and the slight shift observed due to the bond formation between metal and nitrogen of IQL.45,46 Protons in the phenyl ring had singlet, doublet, and multiplet recorded around δ 8.11–6.93 ppm.46,47 A peak at δ 155.92 ppm corresponds to the carbon atom carrying a hydroxyl group, or phenol (=C–OH), in the ligand structure.48 The aromatic carbon’s chemical shift signals of the ligands were recorded in the range of δ 148.52–111.77 ppm.49 From the 13C and 1H NMR spectrum data, the ligand signals were shifted upon metal ion complexation with to the IQL ligand under study.
3.3. FTIR Spectral Analysis of the Ligand and Its Cu(II) Complexes
The FTIR spectra of both IQL and its Cu(II) complexes were analyzed within the range of 4000–400 cm–1 to identify the frequencies associated with functional groups. The results of the vibrational frequencies are presented in Table 2 and Figure 1. Specifically, the absorption peak observed at 1601 cm–1 in the spectrum of IQL indicates the presence of the imine (–C=N–) group.50 The observed peak shifted by 2 to 6 cm–1 in the Cu(II) complexes toward lower or higher wavelength regions. This alteration suggests an active interaction between the nitrogen atom of the azomethine group and the metal ion. Additionally, the phenolic O–H band is present at 3742 cm–1 in the IQL spectrum.51 The observed peak in the complexes undergoes a shift of 14 to 20 cm–1 without deprotonation, indicating coordination of oxygen to the copper metal ion. Furthermore, the stretching frequency of the (–C–O) band in the copper complexes is lower compared to that of IQL, providing strong evidence of coordination between the metal ion and phenolic oxygen. The emergence of three bands in the regions of 586–596 cm–1 and 530–457 cm–1 can be attributed to stretching frequencies of M–O and M–N bonds, respectively, confirming the successful complexation of the copper metal ion with the ligand.52 A new band observed in the complexes at around 3457–3468 cm–1 indicates that water molecules were coordinated with the core metal ion.53,54
Table 2. Selected FTIR Data of IQL and its Cu(II) Complexes.
| Group | IQL | CuL | CuL2 |
|---|---|---|---|
| OH | 3742 | 3726 | 3728 |
| C–O | 1308 | 1334 | 1322 |
| M ← OHR | 3457 | 3468 | |
| sNH2 | 1734 | 1644 | 1647 |
| v(C=N) | 1601 | 1603 | 1606 |
| 1560 | 1569 | 1578 | |
| M ← O | 596 | 591 | |
| M ← N | 530, 457 | 500, 458 |
Figure 1.
FTIR spectra of IQL and its complexes, CuL and CuL2.
3.4. Electronic Spectra of IQL and Its Cu(II) Complexes
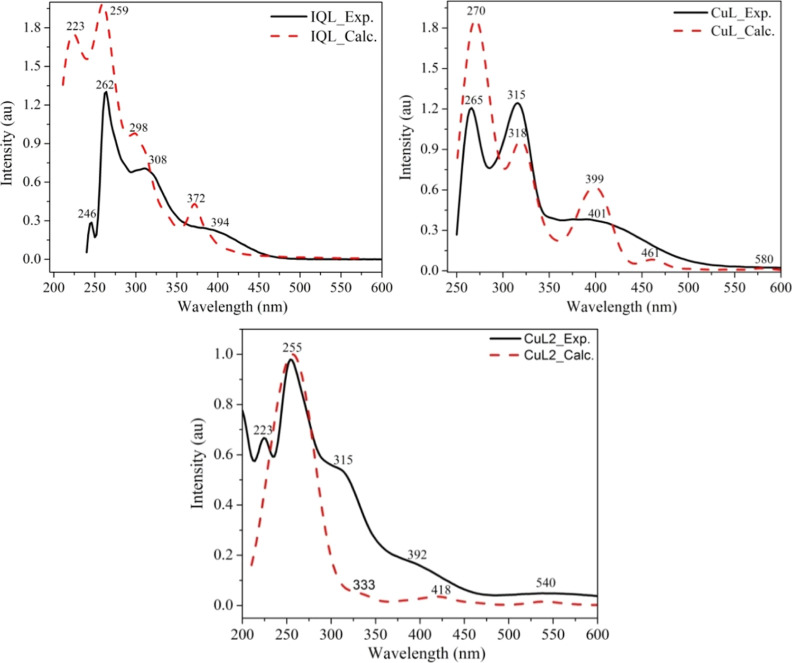
Results from the experimental and TD-DFT absorption studies are summarized in Table S2, Figure 2, and S3. The observed electronic transitions in both experiment and TD-DFT for IQL, occurring approximately at 262/259, 308/298, and 394/372 nm, are attributed to intraligand charge transitions. These transitions are influenced by the energy levels of the orbitals, particularly π → π* and n → π* transitions within the aromatic ring and C=N groups.55,56 The λmax values exhibited slight shifts either toward higher or lower wavelengths, and new bands appeared within the Cu(II) complexes in comparison to the ligand spectra. These findings validate the formation of metal complexes, as indicated in Table S2 and Figure S2.
Figure 2.
Experimental and DFT calculated UV–vis spectra of IQL, CuL, and CuL2 complexes. The calculated spectra were red-shifted by 30 nm for better comparison with the corresponding experimental spectra.
Transitions resulting from the metal ion’s d orbital and intraligand transitions are seen at 592/580 nm (experiment/TD-DFT) for CuL.57 The result allows one to ascertain the degree of fission associated with a certain geometry because of extracted transitions such as d → d (electronic transition), MLCT, and intraligand transitions.58 Transitions like n → π* and π → π* in coordinating ligands have been seen in deep ultraviolet areas. Jahn–Teller’s distortion causes the Cu(II) complexes’ UV–vis spectra to have deformed octahedral geometry. Moreover, transitions at 401/399 nm are linked to d → d (t2g →eg) transitions.57−59 The intramolecular charge transfer bands of π → π* and n → π* transitions of the azomethine group C=N is responsible for the absorption band at 265/270 nm. These values also coincide with those of related Schiff-base compounds.60,61 The same observation has also been noted for the CuL2 complex. Overall, the experimental results were found to be in good agreement with those from the TD-DFT calculations. These results further validate the suggested structures.
3.5. Powder X-ray Diffraction
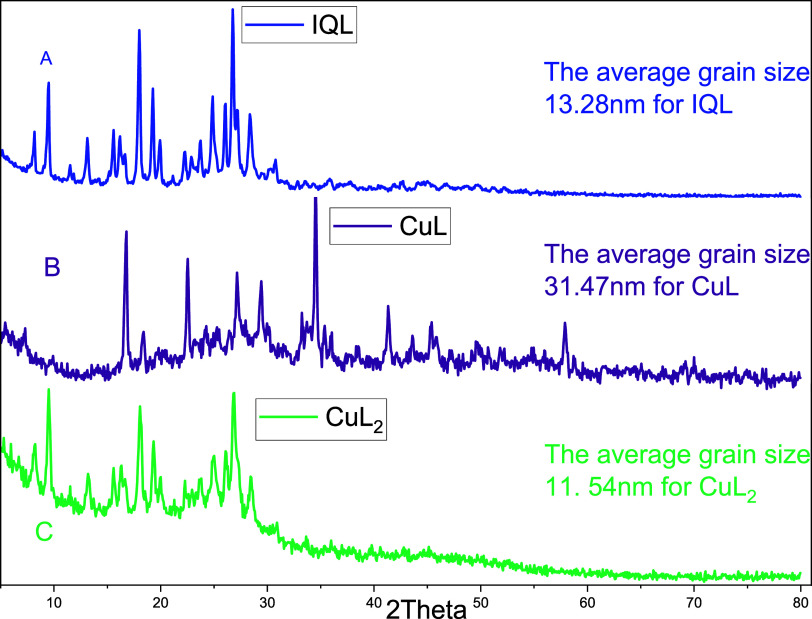
PXRD experiments were carried out on IQL and its Cu(II) complexes at room temperature. The results revealed that the ligand and Cu(II) complexes were all polycrystalline in nature (Figure 3).
Figure 3.
pXRD spectra of the (A) IQL, (B) CuL, and (C) CuL2.
The ligand and its Cu(II) complexes had completely different XRD patterns, confirming the successful complexation of copper metal ions with the ligand. The average grain sizes of the samples were calculated using Scherer’s formula dXRD = 0.9λ/β (cos θ) and the results were found to be 13.28, 31.47, and 11.54 nm for IQL, CuL (1:1), and CuL2 (1:2), respectively. The findings articulate that the complexes were nanocrystalline in size.62 New XRD peaks were observed for CuL at 41.33, 45.41, and 57.91°.63,64 However, the pXRD pattern of CuL2 was similar to that of the ligand, which might be due to masking of the Cu(II) metal ion associated with a higher ratio of the ligand used for the complexation.
3.6. Thermogravimetric Investigation of the Complexes of Copper
Thermal stability assessment of the Cu(II) complexes was executed via TGA across the temperature range of 30 to 1000 °C, employing a heating rate of 10 °C/min under a nitrogen atmosphere. Analysis of the TGA/DTA spectra aimed to discern the presence of water molecules either within or outside the coordination sphere of the Cu(II) complexes. Figure 4 depicts the TGA/DTA spectra of the Cu(II) metal complexes. Results indicated that the copper complexes exhibited thermal stability up to temperatures ranging from 170 to 200 °C. Subsequent to this range, breakdown of the complexes ensued, signifying the presence of water molecules bonded with the metal centers and the onset of the breakdown process. The degradation process of the complexes unfolded in four phases, as elucidated in Figure 4, with the proposed breakdown mechanism outlined in Table 3.
Figure 4.

TGA/DTA of the copper complexes, CuL (1:1) and CuL2 (1:2).
Table 3. Thermal Degradation of the Cu(II) Complexes with Their Suggested Molecules.
| complexes | temperature range (°C) | % removed | proposed degraded molecules |
|---|---|---|---|
| [CuL(H2O)2Cl2] | 200–290 | 10.03 | two water molecules |
| C22H20Cl3CuN3O3S | 290–383 | 16.56 | two chlorine molecules |
| MW: 576.38 g/mol | 383–833 | 62.45 | C22H16ClN3OS the ligand |
| >833 | 10.96 | CuO | |
| [CuL2Cl2] | 170–225 | 8.08 | two chlorine molecules |
| C44H32Cl4CuN6O2S2 | 225–545 | 25.57 | C12H16N2S |
| MW: 946.25 g/mol | 545–721 | 42.41 | C22H16ClN3S |
| 721–903 | 22.07 | C11H7ClO |
From the spectroscopic characterizations, conductivity measurement, and chloride test data, it is concluded that the structure of the Cu(II) complexes has octahedral geometries as illustrated in Scheme 2.
Scheme 2. Scheme of Synthesis of the Copper (II) Complexes and Their Proposed Structures.
3.7. Biological Activity Analysis of the Synthesized Compounds
3.7.1. Antibacterial Activities’ Analysis Result
The ligand and its Cu(II) complexes exhibited antagonistic effects against both Gram-positive and Gram-negative bacteria, as presented in Table 4 and Figure S3. The inhibition zones ranged from 7.33 to 9.01 mm, with the Cu(II) complexes demonstrating significantly higher antagonism compared to the ligand at α = 0.01. Notably, CuL displayed the highest inhibition zone (9.01 mm) against the Gram-negative bacteria P. aeruginosa. The antibacterial activities of CuL were consistent across both Gram-negative and Gram-positive bacterial strains, ranging from 8.04 mm to 9.01 mm (Table 4). CuL and CuL2 exhibited stronger inhibition against Gram-positive bacteria (S. aureus) compared to the Gram-negative bacteria.
Table 4. Antimicrobial Properties of the Ligand and Cu(II) Complexesa.
| zone
inhibition diameter (mm) |
|||
|---|---|---|---|
| Gram-positive bacteria | Gram-negative
bacteria |
||
| compounds (50 μg/mL) | S. aureus | E. coli | P. aeruginosa |
| IQL | 7.47 ± 0.41 | 7.33 ± 0.47 | 7.67 ± 0.47 |
| CuL | 8.67 ± 0.47 | 8.04 ± 0.82 | 9.01 ± 0.82 |
| CuL2 | 8.33 ± 0.47 | 7.33 ± 0.47 | 7.67 ± 0.47 |
| gentamycin (Pos) | 18.33 ± 0.47 | 17.06 ± 0.10 | 17.67 ± 0.47 |
| DMSO (Neg) | ND | ND | ND |
Pos = positive control, Neg = negative control, ND = not detected.
Overall, the antibacterial activity tests revealed that the copper complexes displayed better antibacterial activity than the ligand. Specifically, CuL demonstrated significantly larger inhibition zones against all tested bacterial pathogens compared to CuL2 (Table 4). Previous studies have shown that a 1:2 Cu(II)-to-ligand ratio of 2-((2-hydroxyethyl)amino)quinoline-3-carbaldehyde (H2L) in methanol exhibited decreased antibacterial efficacy against similar microorganisms, consistent with the findings of the current study.65 This may be related to the metal complexes chelation, which links chelation’s capacity to complex passage over a cell membrane.66
3.7.1.1. MIC of IQL and its Cu(II) Complexes
The MIC of the Ligand and CuL were found to be 25 and 12.5 μg/mL, respectively, against the tested bacterial pathogens (Table 5). CuL2 was strong against S. aureus ATCC 25926 compared to the Gram-negative bacterial pathogens tested in this study. Related literature reports on copper complexes also showed stronger MIC against Gram-positive bacteria as compared to Gram-negative bacteria.67
Table 5. Minimum Inhibitory Concentration (mg/mL) of IQL and Its Cu(II) Complexes.
| samples (μg/mL) |
||||
|---|---|---|---|---|
| bacterial strains | IQL | CuL | CuL2 | gentamycin |
| S. aureus ATCC 25926 | 25 | 12.5 | 12.5 | 3.375 |
| E. coli ATCC 25922 | 25 | 12.5 | 25 | 1.688 |
| P. aeruginosa ATCC 248 | 25 | 12.5 | 25 | 1.688 |
However, unlike a previous report,67 the present Cu(II) complexes inhibited Gram-positive bacterial strains at 12.5 μg/mL although the minimum inhibition zone did not coincide. In general, the complexes’ ability to limit bacterial growth was not improved by the integration of ligand and copper metal ions. Previous study also showed that there is a lesser inhibitory potential of the ligands in comparison to a pure metal complex.68
3.7.2. Antioxidant Activity Analysis Result
The capacity of the ligand and its Cu(II) complexes to scavenge 2,2-diphenyl-1-picrylhydrazyl (DPPH) free radicals was evaluated in vitro, with ascorbic acid serving as the standard. This assessment involved measuring the reduction in absorbance of DPPH at 517 nm. Figure 5 and Table S3 illustrate the increase in scavenging activity with rising concentrations of the compounds. In comparison to the reference ascorbic acid, all complexes exhibited moderate activity, with CuL demonstrating superior antioxidant ability compared to CuL2 and IQL. The half-maximal radical scavenging concentration (IC50) of the synthesized compounds was calculated using GraphPad Prism 9.5.1 software, with IQL (239 ± 0.83 μg/mL) > CuL2 (192.5 ± 0.86 μg/mL) > CuL (153.3 ± 1.02 μg/mL). These findings indicate that CuL possesses potent radical scavenging activity compared to the other compounds.
Figure 5.

Antioxidant activities of IQL and its Cu(II) complex.
Compared to the ligand, the copper complexes demonstrate improved radical scavenging activity, as observed through UV–vis measurements of DPPH radical absorbance at 517 nm. The addition of the copper complex leads to a reduction in the visible absorption band at 517 nm, indicating a decrease in the quantity of DPPH radical due to its conversion from DPPH· to DPPH. Prior research has suggested that the antioxidant activity of Cu(II) complexes, such as those involving lidocaine and ibuprofen amide-phenanthroline, is primarily attributed to the acceptance of one electron from the Cu(II) complex.69 As a result, the decreased absorbance is due to the formation of DPPH anion as a function of increased Cu(II) complex concentration.69 Similarly, the band at 517 nm may have disappeared in the excess Cu(II) complex as a result of DPPH switching from its anion form to its DPPH–H form by taking one proton from the metal complex.60,61 The scavenging reaction of the DPPH radical by the Cu(II) complex was proposed to occur in two steps, with the first step being electron-accepting and the second step being proton-accepting as illustrated in Figure 6A.70,71
Figure 6.
UV–vis spectra of Cu(II) and DPPH radical with proposed mechanism (A) and an experimental absorbance of DPPH radical scavenging activities (B).
3.7.3. In Vitro Cytotoxicity Assay of the Ligand and Its Cu(II) Complexes
The human breast cancer cell line (MCF-7) was used to evaluate the in vitro cytotoxicity of the synthesized IQL and its copper complexes (CuL and CuL2) at different doses (6.125–200 μg/mL). Figure 7 shows that CuL inhibited proliferation of MCF-7 breast cancer cells in a dose-dependent manner, with the highest in vitro cytotoxicity observed between 50 and 200 μg/mL. A dose-dependent growth inhibitory effect was observed as the proliferation of MCF-7 cells decreased with an increase in the concentration of synthesized compounds.60,61 All the synthesized compounds showed moderate cytotoxicity against the MCF-7 breast cancer cell line.72CuL demonstrated the highest cytotoxic potency, with the lowest half-maximal inhibitory concentration (IC50) of 43.82 ± 2.351 μg/mL, whereas CuL2 and IQL exhibited higher IC50 values of 132.20 ± 5.687 μg/mL and 117.60 ± 1.098 μg/mL, respectively (Table S2). Although all the synthesized compounds were moderately cytotoxic against MCF-7 cells, their cytotoxic profiles demonstrated less potency compared to cis-platin, which had IC50 values of 5.69 μg/mL.
Figure 7.

Cytotoxicity effect of IQL, CuL, and CuL2 against MCF-7 cancer cell lines.
3.7.4. Quantum Chemical Analysis
Table 6 displays the results for the quantum chemical descriptors, which include the band gap energy, as well as the HOMO and LUMO Eigenvalues. These descriptors have been found to exhibit significant correlations with various biological activities, such as cytotoxicity, antioxidant, and antibacterial properties. For the ligand, the band gap energy (eV) was estimated to be 3.409, whereas 3.255 for CuL2, and 2.062 for CuL. Notably, the metal complexes exhibited a decrease in band gap energy, accompanied by an increase in their global softness, with values of 0.293, 0.307, and 0.485 eV, respectively, for CuL, CuL2, and IQL. These findings underscore the enhanced biological relevance of the synthesized complexes compared to the bare ligand. Consequently, CuL demonstrated significant contributions to its better in vitro cytotoxicity and antioxidant activity.
Table 6. Quantum Chemical Descriptors (in eV) of the Ligand and Its Cu(II) Complexes.
| HOMO | LUMO | Eg | χ | μ | η | σ | ω | Nu | |
|---|---|---|---|---|---|---|---|---|---|
| IQL | –6.02438 | –2.61531 | 3.409 | 4.320 | –4.320 | 1.705 | 0.293 | 5.474 | 0.183 |
| CuL | –5.87308 | –3.81126 | 2.062 | 4.842 | –4.842 | 1.031 | 0.485 | 11.372 | 0.088 |
| CuL2 | –6.42165 | –3.16700 | 3.255 | 4.794 | –4.794 | 1.627 | 0.307 | 7.062 | 0.142 |
3.7.5. Molecular Docking
Tables S5, S6, and 7 present the 3D and 2D interactions of the synthesized IQL and the Cu(II) complexes with residual amino acids from S. aureus, E. coli, and ERα, along with their respective binding energies and inhibition constants. The interaction of IQL with residual amino acids from S. aureus resulted in a binding energy of −11.31 kcal/mol and an inhibition constant of 0.05 μM. A total of 15 amino acid interactions were identified, involving different binding modes: 2 via hydrogen bonds, 9 via van der Waals forces, 2 via π–alkyl interactions, and 1 via π–sigma interaction, as depicted in Figure S5.
Table 7. Molecular Docking Scores and the Corresponding Prominent Residual Amino Acid Interactions of the Ligand and Its Cu(II) Complexes against Estrogen Receptor Alpha (ERα; PDB: 5GS4).
| Cpds | binding energy (kcal/mol) | inhibition constant (Ki) | H-bonding | van der Waals | π–alkyl | π–π | π–anion |
|---|---|---|---|---|---|---|---|
| IQL | –7.63 | 2.53 μM | Glu353, Glu323 | Arg394,Ala322, Asn359,Arg363, Gly390,Ile386, Leu387,Lys449, Phe445,Trp360 | Met357, His356 | ||
| CuL | –8.79 | 0.36 μM | Arg394, Ile386, Glu353 | Glu323,His356, Leu387,Gly390, Leu391 | Met357 | Phe445, Trp393, Trp360 | Lys449 |
| CuL2 | –8.66 | 0.45 μM | Glu323, Asp 321 | Pro406,Gly390, Leu387,Trp393, Phe445,Arg363, Ala322, Trp360 | Met357 | Arg394 | |
| Cisplatin | –6.09 | 34.40 μM | Glu471,Asp374, Ser468 | Thr371 | Lys467 |
The interaction of the CuL with residual amino acids of S. aureus was evaluated, revealing a binding energy of −13.82 kcal/mol and an inhibition constant of 7.5 × 10–5 μM. A total of 18 residual amino acid interactions were identified, occurring through various binding modes: 4 via hydrogen bonds, 10 via van der Waals forces, 3 via π–alkyl interactions, and 1 via π–sigma interaction (Figure S6). Similarly, the CuL2 metal complex exhibited interaction with S. aureus residual amino acids, yielding a binding energy of −12.19 kcal/mol and an inhibition constant of 0.01 μM. A total of 8 residual amino acid interactions were observed, involving different binding modes: 1 via hydrogen bond, 1 via van der Waals forces, 3 via π–alkyl interactions, and 3 via π–sigma interactions (Figure S7). Gentamicin served as a positive control, displaying a binding energy of −10.78 kcal/mol and an inhibition constant of 0.13 μM. A total of 21 residual amino acid interactions were noted, with various binding modes, excluding interactions via π–sigma (Figure S8).
Furthermore, the interactions of IQL, CuL, and CuL2 with residual amino acids of E. coli were analyzed. IQL demonstrated a binding energy of −7.92 kcal/mol and an inhibition constant of 1.57 μM. Sixteen amino acid interactions were identified, involving various binding modes: 2 via hydrogen bonds, 9 via van der Waals forces, 4 via π–alkyl interactions, and 1 via π–sigma interaction (Figure S9). CuL interacted with E. coli residual amino acids, displaying a binding energy of −7.98 kcal/mol and an inhibition constant of 1.41 μM. Fifteen residual amino acid interactions were observed, occurring through different binding modes: 1 via hydrogen bond, 6 via van der Waals forces, 5 via π–alkyl interactions, and 3 via π–sigma interactions (Figure S10). Similarly, CuL2 interacted with E. coli residual amino acids, showing a binding energy of −8.92 kcal/mol and an inhibition constant of 0.29 μM. Eleven residual amino acid interactions were noted, with different binding modes: 3 via hydrogen bonds, 4 via van der Waals forces, 3 via π–alkyl interactions, and 1 via π–sigma interaction (Figure S11). Gentamicin was utilized as a positive control for interaction with E. coli residual amino acids, exhibiting a binding energy of −10.10 kcal/mol and an inhibition constant of 39.31 μM. Sixteen residual amino acid interactions were identified, involving various binding modes, except for interactions via π–sigma (Figure S12).
The docking of IQL, CuL, CuL2, and Cisplatin (used as a positive control) against estrogen receptor alpha was assessed, considering their interaction with residual amino acids through H-bonding, van der Waals, π–alkyl, π–π, and π–anion interaction modes (Table 7 and Figures 8, 9). The interaction of IQL with ERα residual amino acids revealed a binding energy of −7.63 kcal/mol and an inhibition constant of 2.53 μM. A total of 14 amino acid interactions were identified, involving different binding modes: 2 through hydrogen bonds, 10 through van der Waals forces, and 2 through π–alkyl interactions (Figure S13). CuL displayed a binding energy of −8.79 kcal/mol and an inhibition constant of 0.36 μM, with a total of 13 amino acid interactions occurring through various binding modes, including 3 through hydrogen bonds, 5 through van der Waals forces, 1 through π–alkyl, 3 through π–π, and 1 through π–anion interactions (Figure 8). Similarly, CuL2 exhibited a binding energy of −8.66 kcal/mol and an inhibition constant of 0.45 μM, with a total of 14 amino acid interactions occurring through different binding modes: 3 through hydrogen bonds, 9 through van der Waals forces, 1 through π–alkyl, and 2 through π–anion interactions (Figure 9). Cisplatin demonstrated a binding energy and inhibition constant of −6.09 kcal/mol and 34.40 μM, respectively.
Figure 8.
Binding interactions of CuL against estrogen receptor alpha (ERα; PDB: 5GS4).
Figure 9.
Binding interactions of CuL2 against estrogen receptor alpha (ERα; PDB: 5GS4).
4. Conclusions
In conclusion, this study focused on the synthesis and characterization of two Cu(II) complexes, [CuL(H2O)2Cl2] and [CuL2Cl2], with the aim of developing potential therapeutic agents with antimicrobial and cytotoxic properties. Thorough characterization was conducted using various analytical techniques, including FTIR, pXRD, TGA/DTA, UV–vis, TD-DFT, molar conductivity, and chloride test. The synthesized Cu(II) complexes demonstrated thermal stability up to 200 °C, with pXRD confirming their nanocrystalline nature. UV–vis spectra indicated intraligand electron transfer, d–d electron transitions, and ligand-to-metal charge transfer. FTIR confirmed the functional groups of the ligand and complexes, with the complexes found to be nonelectrolytic and chloride-free outside the coordination sphere, suggesting an octahedral geometry. TD-DFT absorption spectra align well with experimental data. Biological evaluations revealed superior antioxidant and antibacterial properties in the Cu(II) complexes compared to IQL, with CuL exhibiting the most potent cytotoxicity, antibacterial, and antioxidant activities. These results were supported by frontier molecular orbital analysis and molecular docking, indicating CuL’s contribution to enhanced cytotoxicity and antioxidant activity. The molecular docking results revealed a higher binding affinity of the Cu(II) complexes to the binding sites of S. aureus compared to the ligand, which is consistent with the experimental findings. This suggests that the CuL complex may have potential therapeutic applications against S. aureus infections. Therefore, it is recommended to conduct further in vivo studies to evaluate the efficacy, safety, and potential therapeutic benefits of the CuL complex in treating S. aureus and MCF-7 breast cancer infections.
Acknowledgments
The authors would like to thank Adama Science and Technology University, Ethiopia, and the University of Botswana for providing research facilities. The authors are also thankful to the Chemistry and Biology Departments, Wollega University, Ethiopia, for providing laboratory space to perform the experimental work, UV–vis spectrophotometry, and antibacterial activity studies.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c02129.
Additional data on the 1H and 13C NMR; UV/vis spectra; molecular docking scores; visualization of the interactions of the synthesized molecules against E. coli, S. aureus, and estrogen receptor alpha; and DFT-optimized geometries (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Ferlay J.; Laversanne M.; Ervik M.; Lam F.; Colombet M.; Mery L.; Piñeros M.; Znaor A.; Soerjomataram I. B. F.. Global Cancer Observatory: Cancer Tomorrow (version 1.1); International Agency for Research on Cancer: Lyon, France, 2024. [Google Scholar]
- Siegel R. L.; Miller K. D.; Wagle N. S.; Jemal A. Cancer statistics, 2023. Ca-Cancer J. Clin. 2023, 73, 17–48. 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- Arnold M.; Morgan E.; Rumgay H.; Mafra A.; Singh D.; Laversanne M.; Vignat J.; Gralow J. R.; Cardoso F.; Siesling S.; et al. Current and future burden of breast cancer : Global statistics for 2020 and 2040. Breast 2022, 66 (September), 15–23. 10.1016/j.breast.2022.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H.; Ferlay J.; Siegel R. L.; Laversanne M.; Soerjomataram I.; Jemal A.; Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Ca-Cancer J. Clin. 2021, 71 (3), 209–249. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Lucaciu R. L.; Hangan A. C.; Sevastre B.; Oprean L. S. Metallo-Drugs in Cancer Therapy: Past, Present and Future. Molecules 2022, 27 (19), 6485–6530. 10.3390/molecules27196485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Xu C.; Gao X.; Yao Q. Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics 2022, 12 (5), 2115–2132. 10.7150/thno.69424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans A.; Kavanagh K. A. Evaluation of metal-based antimicrobial compounds for the treatment of bacterial pathogens. J. Med. Microbiol. 2021, 70 (5), 1–18. 10.1099/jmm.0.001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2019 Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline; World Health Organization: Geneva, 2019. Licence: CC BY-NC-SA 3.0 IGO.; 2019. [Google Scholar]
- ChunYan Z.; RuJian Y.; LiQiang W.; HaiYan H.; JinTao W.; XiangWen L.; XueMin D.; YanShi X. Design, synthesis, and evaluation of aryl-thioether ruthenium polypyridine complexes: A multi-target antimicrobial agents against gram-positive bacteria. Eur. J. Med. Chem. 2022, 240, 114562. 10.1016/j.ejmech.2022.114562. [DOI] [PubMed] [Google Scholar]
- Anthony E. J.; Bolitho E. M.; Bridgewater H. E.; Carter O. W. L.; Donnelly J. M.; Imberti C.; Lant E. C.; Lermyte F.; Needham R. J.; Palau M.; et al. Metallodrugs are unique: opportunities and challenges of discovery and development. R. Soc. Chem. 2020, 11 (48), 12888–12917. 10.1039/d0sc04082g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurca T.; Marian E.; Graţiela L.; et al. Metal Complexes of Pharmaceutical Substances. In Spectroscopic Analyses - Developments and Applications Chemistry; BoD, 2017; pp 123–142. [Google Scholar]
- Claudel M.; Schwarte J. V.; Fromm K. M. New Antimicrobial Strategies Based on Metal Complexes. Chemistry 2020, 2, 849–899. 10.3390/chemistry2040056. [DOI] [Google Scholar]
- Todorov L.; Kostova I. Recent Trends in the Development of Novel Metal-Based. Mol. Pharm. 2023, 28, 1959. 10.3390/molecules28041959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamberi T.; Hanif M. Metal-Based Complexes in Cancer Treatment. Biomedicines 2022, 10 (10), 2573–2578. 10.3390/biomedicines10102573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D.; Zhao L.; Shi X.; Ma X.; Chen Z. Copper in cancer : From pathogenesis to therapy. Biomed. Pharmacother. 2023, 163 (April), 114791–114817. 10.1016/j.biopha.2023.114791. [DOI] [PubMed] [Google Scholar]
- Hong Y.; Boiti A.; Vallone D.; Foulkes N. S. Reactive Oxygen Species Signaling and Oxidative Stress : Transcriptional Regulation and Evolution. Antioxidants 2024, 13 (3), 312–324. 10.3390/antiox13030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Han X.; Zhang T.; Tian K.; Li Z.; Luo F. Reactive oxygen species (ROS) scavenging biomaterials for anti - inflammatory diseases : from mechanism to therapy. J. Hematol. Oncol. 2023, 16 (1), 116–134. 10.1186/s13045-023-01512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aishajiang R.; Liu Z.; Wang T.; Zhou L.; Yu D. Recent Advances in Cancer Therapeutic Copper-Based. Molecules 2023, 28, 2303. 10.3390/molecules28052303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro A. C.; Nunes I. J.; Ferreira W. V.; Tomasini P. P.; Trindade C.; Martins C. C.; Wilhelm E. A.; Oliboni R. d. S.; Netz P. A.; Stieler R.; et al. Antioxidant and Anticancer Potential of the New Cu(II) Complexes Bearing Imine-Phenolate Ligands with Pendant Amine N-Donor Groups. Pharmaceutics 2023, 15 (2), 376. 10.3390/pharmaceutics15020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang X. F.; Morris-Natschke S. L.; Liu Y. Q.; et al. Biologically active quinoline and quinazoline alkaloids part I. Med. Res. Rev. 2018, 38 (3), 775–828. 10.1002/med.21466.Biologically. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella A.; Tanwar O. P.; Saha R.; Ali M. R.; Srivastava S.; Akhter M.; Shaquiquzzaman M.; Alam M. M. Quinoline : A versatile heterocyclic. Saudi Pharm. J. 2013, 21 (1), 1–12. 10.1016/j.jsps.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R.; Kumar K. Synthetic and medicinal perspective of quinolines as antiviral agents. Eur. J. Med. Chem. 2021, 215 (January), 113220–113240. 10.1016/j.ejmech.2021.113220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajani O. O.; Iyaye K. T.; Ademosun O. T. Recent advances in chemistry and therapeutic potential of functionalized quinoline motifs - a review. RSC Adv. 2022, 12 (29), 18594–18614. 10.1039/D2RA02896D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakir M.; Hanif S.; Sherwani M. A.; Mohammad O.; Al-Resayes S. I. Pharmacologically significant complexes of Mn(II), Co(II), Ni(II), Cu(II) and Zn(II) of novel Schiff base ligand, (E)-N-(furan-2-yl methylene) quinolin-8-amine: Synthesis, spectral, XRD, SEM, antimicrobial, antioxidant and in vitro cytotoxic studies. J. Mol. Struct. 2015, 1092, 143–159. 10.1016/j.molstruc.2015.03.012. [DOI] [Google Scholar]
- Ibrahim S.; Naik N.; Shivamallu C.; Raghavendra H.; Shati A. A.; Alfaifi M. Y.; Elbehairi S. E. I.; Amachawadi R. G.; Kollur S. P. Synthesis, structure, and in vitro biological studies of benzothiazole based Schiff base ligand and its binary and ternary Co(III) and Ni(II) complexes. Inorg. Chim. Acta 2024, 559 (August 2023), 121792. 10.1016/j.ica.2023.121792. [DOI] [Google Scholar]
- Dinku D.; Demissie T. B.; Beas I. N.; Eswaramoorthy R.; Abdi B.; Desalegn T. Antimicrobial activities and docking studies of new Schiff base ligand and its Cu (II), Zn (II) and Ni (II) Complexes : Synthesis and Characterization. Inorg. Chem. Commun. 2024, 160, 111903. 10.1016/j.inoche.2023.111903. [DOI] [Google Scholar]
- Shumi G.; Demissie T. B.; Eswaramoorthy R.; Bogale R. F.; Kenasa G.; Desalegn T. Biosynthesis of Silver Nanoparticles Functionalized with Histidine and Phenylalanine Amino Acids for Potential Antioxidant and Antibacterial Activities. ACS Omega 2023, 8, 24371–24386. 10.1021/acsomega.3c01910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries R. M.; Abbott A. N.; Hindler J. A. Understanding and Addressing CLSI Breakpoint Revisions - A Primer for Clinical Laboratories. J. Clin. Microbiol. 2019, 57 (6), 1–36. 10.1128/JCM.00203-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M. P.; Rath S.; Swain S. S.; Ghosh G.; Das D.; Padhy R. N. In vitro antibacterial activity of crude extracts of 9 selected medicinal plants against UTI causing MDR bacteria. J. King Saud Univ., Sci. 2017, 29 (1), 84–95. 10.1016/j.jksus.2015.05.007. [DOI] [Google Scholar]
- El Allouchea Y.; Zaitana H.; Bouachrineb M.; Khalila F. QSAR Modeling and Molecular Docking Studies of 3,5-Disubstituted Indole Derivatives as Pim1 Inhibitors for Combating Hematological Cancer. Phys. Chem. Res. 2024, 12 (3), 631–645. 10.22036/pcr.2023.417665.2425. [DOI] [Google Scholar]
- Ordog R.; Grolmusz V.. Evaluating Genetic Algorithms in Protein-Ligand Evaluating Genetic Algorithms in Protein-Ligand Docking; Springer, 2008; pp 402–413. [Google Scholar]
- Alem M. B.; Damena T.; Desalegn T.; Koobotse M.; Eswaramoorthy R.; Ngwira K. J.; Ombito J. O.; Zachariah M.; Demissie T. B. Cytotoxic mixed-ligand complexes of Cu (II): A combined experimental and computational study. Front. Chem. 2022, 10, 1028957. 10.3389/fchem.2022.1028957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M.; Zhao H.; Liu Q.; et al. Structural Basis of Inhibition of ER # -Coactivator Interaction by Structural Basis of Inhibition of ERα-Coactivator Interaction by High- affinity N-terminus Isoaspartic Acid Tethered Helical Peptides. J. Med. Chem. 2017, 60, 8731–8740. 10.1021/acs.jmedchem.7b00732. [DOI] [PubMed] [Google Scholar]
- Becke A. D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98 (7), 5648–5652. 10.1063/1.464913. [DOI] [Google Scholar]
- Lee C.; Yang W.; Parr R. G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37 (2), 785–789. 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- Stephens P. J.; Devlin F. J.; Chabalowski C. F.; Frisch M. J. Ab Initio Calculation of Vibrational Absorption and Circular Dichroism Spectra Using Density Functional Force Fields. J. Phys. Chem. 1994, 98 (45), 11623–11627. 10.1021/j100096a001. [DOI] [Google Scholar]
- Krishnan R.; Binkley J. S.; Seeger R.; Pople J. A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 1980, 72 (1), 650–654. 10.1063/1.438955. [DOI] [Google Scholar]
- Hay P. J.; Wadt W. R. Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg Ab Initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg. J. Chem. Phys. 1985, 82 (1), 270–283. 10.1063/1.448799. [DOI] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.. et al. Gaussian 16, Revis B01.; Gaussian, Inc.: Wallingford CT, 2016. [Google Scholar]
- Grimme S.; Furche F.; Ahlrichs R. An improved method for density functional calculations of the frequency-dependent optical rotation. Chem. Phys. Lett. 2002, 361 (3–4), 321–328. 10.1016/s0009-2614(02)00975-2. [DOI] [Google Scholar]
- Ismael M.; Abdel-Mawgoud A. M. M.; Rabia M. K.; Abdou A. Design and Synthesis of Three Fe(III) Mixed-ligand Complexes: Exploration of Their Biological and Phenoxazinone Synthase-Like Activities. Inorg. Chim. Acta 2020, 505, 119443. 10.1016/j.ica.2020.119443. [DOI] [Google Scholar]
- Bitew M.; Desalegn T.; Demissie T. B.; Belayneh A.; Endale M.; Eswaramoorthy R. Pharmacokinetics and drug-likeness of antidiabetic flavonoids : Molecular docking and DFT study. PLoS One 2021, 16, 02608533. 10.1371/journal.pone.0260853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed H. M.; Ramadan M. A.; Salem H. H.; Fayed M. A. A. Phytoconstituent Isolation and Cytotoxicity Evaluation of the Egyptian Cassia occidentalis L. Possessing Selective Activity against Lung Carcinoma. J. Chem. 2023, 2023, 1–11. 10.1155/2023/6111058. [DOI] [Google Scholar]
- Wang C.; Fan L.; Pan Z.; Fan S.; Shi L.; Li X.; Zhao J.; Wu L.; Yang G.; Xu C. Synthesis of Novel Indole Schiff Base Compounds and Their Antifungal Activities. Molecules 2022, 27 (20), 6858–6915. 10.3390/molecules27206858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimsza J. M.; Sorte E. G.; Alam T. M. Hydration and Hydroxylation of MgO in Solution: NMR Identification of Proton-Containing Intermediate Phases. ACS Omega 2019, 4 (1), 1033–1044. 10.1021/acsomega.8b02705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Obaidy S. S. M.; Al-Obaidy S. S. M.; Al-Khafaji Y. F. Synthesis, Spectral Analysis, Stability, Antibacterial, and Antioxidant of Fe(II) Mixed Ligands Complex of Imidazole and 1,10-Phenanthroline. J. Med. Chem. Sci. 2022, 5 (7), 1231–1241. 10.26655/JMCHEMSCI.2022.7.12. [DOI] [Google Scholar]
- Hernández W.; Carrasco F.; Vaisberg A.; Spodine E.; Manzur J.; Icker M.; Krautscheid H.; Beyer L. Synthesis, Spectroscopic Characterization, Structural Studies, and in Vitro Antitumor Activities of Pyridine-3-carbaldehyde Thiosemicarbazone Derivatives. J. Chem. 2020, 2020, 1–12. 10.1155/2020/2960165. [DOI] [Google Scholar]
- Khalil E.; Mahmoud W. H.; El Desssouky M. M. I.; Mohamed G. G. Synthesis, Spectral, Thermal and Biological Studies of Some Transition and Inner Transition Schiff base Metal Complexes. Egypt. J. Chem. 2021, 64 (7), 3555–3571. 10.21608/ejchem.2021.71817.3578. [DOI] [Google Scholar]
- Gunawan R.; Nandiyanto A. B. D.; Nandiyanto D. How to Read and Interpret 1H-NMR and 13C-NMR Spectrums. Indones. J. Sci. Technol. 2021, 6 (2), 267–298. 10.17509/ijost.v6i2.34189. [DOI] [Google Scholar]
- Mahdy A.; Mohamed M. G.; Aly K. I.; Ahmad H. B.; Emam H. E. Liquid crystalline polybenzoxazines for manufacturing of technical textiles: Water repellency and ultraviolet shielding. Polym. Test. 2023, 119, 107933. 10.1016/j.polymertesting.2023.107933. [DOI] [Google Scholar]
- Mohammad S.; Ahmed S.; Badawi A.; El-Desouki D. Activated Carbon Derived from Egyptian Banana Peels for Removal of Cadmium from Water. J. Appl. Life Sci. Int. 2015, 3 (2), 77–88. 10.9734/JALSI/2015/16652. [DOI] [Google Scholar]
- Aljazzar S. Composition and Decomposition of Several Schiff Base Metal Complexes Containing Co (II) and Ni (II) Ions : Spectroscopic Analysis. Moroccan J. Chem. 2024, 12 (1), 1–448. 10.48317/IMIST.PRSM/morjchem-v12i1. [DOI] [Google Scholar]
- Al-daffay R. K. H.; Al-Hamdani A. A. S. Synthesis, Characterization, and Thermal Analysis of a New Acidicazo Ligand ’ s Metal Complexes Abstract . Baghdad Sci. J. 2023, 20 (1), 0121. 10.21123/bsj.2022.6709. [DOI] [Google Scholar]
- Khalaf M. M.; El-lateef H. M. A.; Gouda M.; Sayed F. N. Design, Structural Inspection and Bio-Medicinal Applications of Some Novel Imine Metal Complexes Based on Acetylferrocene. Materials 2022, 15, 4842. 10.3390/ma15144842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolková R.; Smolko L.; And M. P.; Samol’ová E. Pentacoordinate Mixed-Ligand Zn (II) Complexes with Flufenamato and Niflumato Ligands: Crystal Structure, Spectral Characterization and Biological Properties. Crystals 2023, 13 (10), 1460. 10.3390/cryst13101460Academic. [DOI] [Google Scholar]
- Eichhorn T.; Dimic D.; Markovic Z.; Kaludjerovic G. Synthesis, spectroscopic characterization and DFT analysis of dichlorido (η 6 - p -cymene) ruthenium (II) complexes with isonicotinate-polyethylene glycol ester ligands. J. Serbian Chem. Soc. 2023, 88 (12), 1335–1354. 10.2298/jsc230412070e. [DOI] [Google Scholar]
- Abu-Dief A.; El-dabea T.; El-Khatib R.; Abdou A.; El-Remailya M. A. E. A. Development of New Mixed Cu(II) Chelate Based on 2-Benzimidazolylguanidine and Phenanthroline Ligands: Structural Elucidation, Biological Evaluation, DFT and Docking Approaches. Sohag J. Sci. 2024, 9 (2), 174–185. 10.21608/sjsci.2023.235198.1122. [DOI] [Google Scholar]
- Kumar S. V.; Kumar S. P.; Rupesh D.; Nitin K. Immunomodulatory effects of some traditional medicinal plants. J. Chem. Pharm. Res. 2011, 3 (1), 675–684. [Google Scholar]
- Abbas A. M.; Nasrallah H. H.; Aboelmagd A.; Kishk S. M.; Boyd W. C.; Kalil H.; Orabi A. S. Design, Synthesis, Anti-Inflammatory Activity, DFT Modeling and Docking Study of New Ibuprofen Derivatives. Int. J. Mol. Sci. 2024, 25 (6), 3558. 10.3390/ijms25063558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. Z.; Alimuddin K. S. A.; Khan S. A. A Review on Schiff Base as a Versatile Fluorescent Chemo-Sensors Tool for Detection of Cu2+ and Fe3+ Metal Ion. J. Fluoresc. 2023, 33, 1241–1272. 10.1007/s10895-022-03102-1. [DOI] [PubMed] [Google Scholar]
- Manvatkar V. D.; Patle R. Y.; Meshram P. H.; Dongre R. S. Azomethine-functionalized organic–inorganic framework: an overview. Chem. Pap. 2023, 77 (10), 5641–5662. 10.1007/s11696-023-02889-y. [DOI] [Google Scholar]
- Ejidike I. P. Cu(II) Complexes of 4-[(1E)-N-{2-[(Z)-benzylidene-amino]ethyl} ethanimidoyl]benzene-1,3-diol schiff base: Synthesis, spectroscopic, in-vitro antioxidant, antifungal and antibacterial studies. Molecules 2018, 23 (7), 1581–1618. 10.3390/molecules23071581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoorazm M.; Moradi P.; Noori N.; Azadi G. l-Arginine complex of copper on modified core–shell magnetic nanoparticles as reusable and organic–inorganic hybrid nanocatalyst for the chemoselective oxidation of organosulfur compounds. J. Iran Chem. Soc. 2021, 18 (2), 467–478. 10.1007/s13738-020-02040-8. [DOI] [Google Scholar]
- Khakhlary P.; Baruah J. B. Studies on cluster, salt and molecular complex of zinc-quinolinate. J. Chem. Sci. 2015, 127 (2), 215–223. 10.1007/s12039-015-0781-6. [DOI] [Google Scholar]
- Damena T.; Alem M. B.; Zeleke D.; Desalegn T.; Eswaramoorthy R.; Demissie T. B. Synthesis, characterization, and biological activities of zinc(II), copper(II) and nickel(II) complexes of an aminoquinoline derivative. Front. Chem. 2022, 10 (November), 1–21. 10.3389/fchem.2022.1053532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebout O.; Trifa C.; Bouacida S.; Boudraa M.; Imane H.; Merzougui M.; Mazouz W.; Ouari K.; Boudaren C.; Merazig H. Two new copper (II) complexes with sulfanilamide as ligand : Synthesis, structural, thermal analysis, electrochemical studies and antibacterial activity. J. Mol. Struct. 2022, 1248, 131446. 10.1016/j.molstruc.2021.131446. [DOI] [Google Scholar]
- Alem M. B.; Desalegn T.; Damena T.; Alemayehu Bayle E.; Koobotse M. O.; Ngwira K. J.; Ombito J. O.; Zachariah M.; Demissie T. B. Cytotoxicity and Antibacterial Potentials of Mixed Ligand Cu(II) and Zn(II) Complexes: A Combined Experimental and Computational Study. ACS Omega 2023, 8 (14), 13421–13434. 10.1021/acsomega.3c00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Qadsy I.; Saeed W. S.; Al-Odayni A. B.; Alrabie A.; Al-Faqeeh L. A. S.; Al-Adhreai A.; Al-Owais A. A.; Semlali A.; Farooqui M. Antidiabetic, antioxidant and cytotoxicity activities of ortho- and para-substituted Schiff bases derived from metformin hydrochloride: Validation by molecular docking and in silico ADME studies. Open Chem. 2023, 21 (1), 1–15. 10.1515/chem-2023-0125. [DOI] [Google Scholar]
- Tabrizi L.; Dao D. Q. A.; Vu T. A. Experimental and theoretical evaluation on the antioxidant activity of a copper (II) complex based on lidocaine and ibuprofen amide-phenanthroline agents. RSC Adv. 2019, 9, 3320–3335. 10.1039/c8ra09763a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibi Sadeer N.; Montesano D.; Albrizio S.; Zengin G.; Mahomoodally M. F. The Versatility of Antioxidant Assays in Food Science and Safety — Chemistry, Applications, Strengths, and Limitations. Antioxidants 2020, 9 (8), 709. 10.3390/antiox9080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurgul I.; Hricov J.; Mazuryk O.; Hricov Z.; Brindell M. Enhancement of the Cytotoxicity of Quinazolinone Schiff Base Derivatives with Copper Coordination. Inorganics 2023, 11, 391. 10.3390/inorganics11100391. [DOI] [Google Scholar]
- González-Larraza P. G.; López-Goerne T. M.; Padilla-Godínez F. J.; González-López M. A.; Hamdan-Partida A.; Gómez E. IC50Evaluation of Platinum Nanocatalysts for Cancer Treatment in Fibroblast, HeLa, and DU-145 Cell Lines. ACS Omega 2020, 5 (39), 25381–25389. 10.1021/acsomega.0c03759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.